Based on Network Pharmacology, Molecular Docking and Experiments to Explore the Anti-gastric Cancer Effect of Apigenin
-
摘要: 目的:基于网络药理学与分子对接技术结合CCK8法、细胞划痕、细胞侵袭、平板克隆形成实验探讨芹菜素治疗胃癌作用。方法:本文通过ChEMBL、UniProt、GeneCards等数据库及jvenn获得芹菜素治疗胃癌的潜在靶点,结合蛋白质相互作用(PPI)分析,基因本体(GO)富集分析、京都基因与基因组百科全书(KEGG)通路富集分析进行作用机制探讨,进一步采用Auto dock软件进行分子对接,最后通过细胞增殖与毒性实验(Cell counting kit-8,CCK8)验证芹菜素对胃癌细胞的抑制作用。结果:芹菜素治疗胃癌潜在作用靶点为52个;GO分析表明,芹菜素抗胃癌机制可能与调节信号传导、化学突触传递、蛋白水解等有关;KEGG分析发现其通过调节神经活性配体-受体相互作用、氮代谢等信号通路发挥抗癌作用;分子对接结果显示配体和受体的结合能为ACHE|−9.1|>MAOA|−8.8|>CA2|−7.4|>DPP4|−7.3|>CA1|−7.2|=GRM5|−7.2|>ADA|−6.8|>CASP3|−5.8|,表明配体和受体具有良好的结合性。体外实验表明,芹菜素对胃癌细胞系有明显的抑制作用,其可能是通过降低MAOA、DPP4、AChE的表达,升高CA2的表达对胃癌起到抑制作用。结论:芹菜素具有抗胃癌的作用,其发挥作用涉及到的主要靶点有AChE、CA1、CA2、CASP3、ADA、MAOA、DPP4、GRM5,展现了芹菜素多成分、多靶点、多途径的优点,为后续研究芹菜素抗胃癌作用提供参考。Abstract: Objective: Network pharmacology, molecular docking technology, and CCK8, wound healing, cell invasion, plate colony formation assay were used to explore the effect of apigenin in the treatment of gastric cancer. Methods: The ChEMBL, UniProt, and GeneCards databases and jvenn tools were used to obtain the potential anticancer targets of apigenin. Then, the PPI network analysis, gene ontology (GO) enrichment analysis and Kyoto Encyclopedia of genes and genomes (KEGG) pathway enrichment analysis of key targets were used to investigate the mechanism, and Auto dock software was used for further molecular docking. Lastly, the inhibitory effect of apigenin on gastric cancer cells was verified through cell counting kit-8 (CCK8) experiments. Results: There were 52 potential targets of apigenin in the treatment of gastric cancer. GO analysis showed that apigenin might exert its anti-gastric cancer mechanism by regulating signal transduction, chemical synaptic transmission, and proteolysis. KEGG analysis showed that apigenin played a role by regulating the neural active ligand receptor interaction, nitrogen metabolism and other signaling pathways. The molecular docking results showed that the binding energy of ligand and receptor was: AChE|−9.1|>MAOA|−8.8|>CA2|−7.4|>DPP4|−7.3|>CA1|−7.2|=GRM5|−7.2|>ADA|−6.8|>CASP3|−5.8|, indicating that the ligand and receptor had good binding affinity. In vitro tests exhibited that apigenin possessed significant inhibitory effect on gastric cancer cell lines, which might be due to its ability to reducing the expression of MAOA, DPP4, AChE and increasing the expression of CA2. Conclusion: Apigenin has the effect of anti-gastric cancer, and the main targets involved in its effect are AChE, CA1, CA2, CASP3, ADA, MAOA, DPP4, GRM5, which shows the advantages of apigenin in multi-component, multi-target, and multi-channel, and provides a reference for the follow-up study of the anti-gastric cancer effect of apigenin.
-
Keywords:
- network pharmacology /
- molecular docking /
- apigenin /
- gastric cancer
-
胃癌(Gastric cancer,GC)是一种源于胃黏膜上皮的恶性肿瘤,在我国各种恶性肿瘤中发病率居首位[1],目前,手术和化疗是常用的治疗方法,但在临床治疗的过程中,大多数抗癌药物具有严重的不良反应,长时间使用会产生一定的耐药性,甚至会发生并发症。因此,研发副作用小,治疗效果好的新型抗胃癌药物具有重要的现实意义。目前,从天然植物中筛选出特异性强的新型靶向抗肿瘤药物,已成为研究开发抗肿瘤药物的重要途径。
芹菜素(Apigenin)为4,5,7—三羟基黄酮,又称为芹黄素、洋芹素,是一种来源于伞型科植物的小分子可食用[2]黄酮类化合物,纯品为黄色晶体状粉末[3]。常见的含有芹菜素的果蔬有猕猴桃、葡萄柚、金桔、芹菜、香菜、甘蓝、薄荷等,其中香菜中的芹菜素是芹菜中的180倍[4−5]。芹菜素不仅存在于水果和蔬菜中,还存在于各类中草药中,如蒲公英[6]、洋甘菊[7]、芫花[8]。据文献报道,芹菜素具有降糖[9]、降压[10]、抗炎[11]、抗氧化[12]、抑菌[13]等功效。目前,研究发现,芹菜素能够调控多种肿瘤细胞的生物行为,能够阻滞细胞周期、促进细胞凋亡,抑制肿瘤细胞的侵袭与迁移,且具有毒性小、致突变作用低等优势[5−6]。林沈国等[14]研究表明芹菜素可以通过抑制细胞增殖、凋亡和分化来抑制乳腺癌。崔昭阳等[15]用芹菜素对人胃癌细胞SGC-7901做了研究,芹菜素通过激活死亡受体参与诱导并对胃癌有一定的治疗作用。崔迎红等[16]研究了芹菜素对肝癌的作用。目前芹菜素在胃癌中的作用机制研究鲜有报道。
网络药理学作为药理学的一个新领域,是基于主成分通过系统生物学理论构建疾病生物靶点和药物成分之间的网络联系,从而获得治疗疾病的有效成分[17],是一门基于系统生物学理论、生物系统网络分析和多目标药物分子设计特定信号节点选择的新学科。中医药配方的作用机制具有多个目标和水平的特点。该机制类似于网络药理学的完整性、系统化和全面性,因此网络药理学适合研究中药化合物的药理学机制[18]。
鉴于此,本研究采用网络药理学与分子对接筛选活性成分芹菜素抗胃癌的靶点,对抗人胃癌细胞(AGS、MGC-803)的生物过程与分子机制进行全面阐述并进一步挖掘抗胃癌的关键基因,为以后研究芹菜素提供新的思路和理论数据。
1. 材料与方法
1.1 材料与仪器
人胃癌细胞系AGS、MGC-803 湖州市中心医院细胞库;芹菜素标准品(分析纯) 上海阿拉丁生化科技公司;RPMI-1640培养液、胎牛血清(Fetal bovine serum,FBS) 美国Hyclone公司;磷酸盐缓冲溶液(Phosphate buffered saline,PBS) 白鲨生物科技公司;0.25%胰酶、青-链霉素双抗(Penicillin-streptomycin,PS) 美国Gibco公司;CCK-8试剂盒、异丙醇、氯仿、多聚甲醛、结晶紫染料 南京诺唯赞生物科技公司;Trizol 福州科净生物科技有限公司;Transwell细胞小室 耐思生物科技公司。
Heracell Vios 160i CR二氧化碳培养箱、Multiskan FC多功能酶标仪、Sorvall LYNX 6000高速冷冻离心机、ABI7500荧光定量PCR仪 赛默飞世尔科技(中国)有限公司;Primo Vert倒置相差显微镜 德国蔡司公司。
1.2 实验方法
1.2.1 数据库平台
通过网络药理学分析芹菜素抗胃癌的作用,采用的数据库及网址见表1。
表 1 网络药理学分析的参照数据库及网址Table 1. Database and website for network pharmacology analysis数据库 名称 网址 ChEMBL 药物化学数据库 https://www.ebi.ac.uk/chembl/ GeneCards 人类基因综合数据库 https://www.genecards.org UniProt 蛋白质信息数据库 https://www.uniprot.org jvenn 在线韦恩图 https://jvenn.toulouse.inrae.fr/ STRING 蛋白互作数据库 https://cn.string-db.org David 注释、可视化和综合发现数据库 https://david.ncifcrf.gov PDB 国际蛋白质结构数据库 https://www.rcsb.org Bioinformatics 微生信数据平台 http://www.bioinformatics.com.cn 1.2.2 芹菜素及胃癌相关靶点的预测
以“Apigenin”为关键词在ChEMBL数据库中搜索靶蛋白信息,在UniProt数据库中将找到的靶点统一转化为基因名。将相关信息导出建立芹菜素靶蛋白列表。以“Gastric cancer”为关键词检索GeneCards数据库,将获得的基因名称等相关信息导出建立胃癌靶基因列表,用于后续分析。
1.2.3 PPI网络的构建及构建成分网络靶点图
将1.2.2中得到的芹菜素与胃癌相关靶点输入到jvenn中,获得二者的交集靶点。将共同的靶点输入到STRING数据库中,选择“Homo sapiens”,设置相互信值0.400,隐藏游离的靶点,得到PPI网络。把下载得到的结果导入到Cytoscape 3.9.1软件中,构建芹菜素与胃癌的网络关系图,并设置between来选取核心靶点,并调整靶点位置使之更清晰。
1.2.4 GO功能富集和KEGG通路富集分析
将所收集到的交叉靶点导入到David数据库中,分别得到生物过程(Biological process,BP)、细胞组成(Cellular component,CC)、分子功能(Molecular function,MF)及信号通路的靶点信息。将这些数据下载并选择前25个核心靶点,导入到微生信平台,通过GO富集分析绘制气泡图和信号通路。
1.2.5 分子对接
1.2.3中得到的核心靶点前八位,在PDB数据库中查找靶蛋白的pdb格式,导入至Pymol软件中进行去除小分子和水的预处理,后导入至Autodock Tools中加氢,导出为pdbqt格式。在Chem数据库中搜索并下载芹菜素的3D结构为mol2格式,在Pymol软件中导出为pdbqt格式。使用Autodock Vina将二者进行对接,取结合能绝对值最高的并用Pymol做可视化分析。
1.2.6 体外实验
细胞株用含有10%的RPMI-1640培养液,置于37 ℃、5% CO2,饱和湿度的培养箱中培养。
1.2.6.1 CCK-8法检测细胞活力
取细胞汇合度达到80%对数生长期的胃癌细胞(AGS,MGC-803),使用细胞计数仪计数,细胞计数后,以4×104/孔的密度接种于96孔板中(100 µL/孔),96孔板边缘一圈加入PBS,避免受到蒸发影响结果;在培养箱中培养24 h后,弃去旧培养基,加入10%血清的1640培养液培养,培养箱条件为37 ℃、5% CO2,孵育4 h。空白对照组:加10%血清的1640完全培养基;实验组:不同浓度的芹菜素标准液(1、2、4、8、16、32 µmol/L),培养时间为24 h;24 h后从培养箱中取出96孔板,并向其中加入CCK-8试剂(0.5 mg/mL)100 µL,继续孵育2~4 h,颜色明显变化后,测450 nm处的吸光度。
细胞生存率(%)=(As−AbAc−Ab)×100 式中:As为实验孔吸光度(含细胞、培养液、CCK-8溶液和药物溶液);Ac为对照孔吸光度(含细胞、培养液、CCK-8溶液,不含药物);Ab为空白孔吸光度(含培养液、CCK-8溶液,不含细胞、药物)。
1.2.6.2 细胞划痕法检测细胞愈合能力
取对数生长期的胃癌细胞(AGS,MGC-803),以每孔5×105接种于6孔板中,37 ℃、5% CO2下培养24 h;24 h后,吸去培养基,在细胞培养皿上制造划痕,同时设置无芹菜素溶液的完全培养基为对照组,以及不同浓度芹菜素溶液的培养基组,使用8 µmol/L和16 µmol/L的芹菜素分别干预AGS和MGC-803细胞,经24 h给药培养并划痕,分别在0、24 h拍照进行比较。设计三组重复实验。
1.2.6.3 Transwell实验检测细胞侵袭能力
将处于对数生长期的细胞,吸去培养基,加磷酸缓冲盐溶液清洗,加胰酶消化,加入1640培养液终止消化,200×g离心5 min;弃去上清,加入1640培养液重悬细胞,进行细胞计数,约每孔5×105;在12孔板中加入500 µL含10% FBS的1640培养液,放入Transwell细胞小室,上室加入200 µL细胞悬液,静置15 min,避免细胞脱落,沉降。静置后再往上室中分别加入8 µmol/L和16 µmol/L芹菜素的无血清培养液,再将12孔板轻放入培养箱中培养24 h;24 h后,取出Transwell小室吸走培养液,PBS洗三次。在12孔板的干净孔中加入4%多聚甲醛2 mL,小室底面浸入溶液中固定10~15 min;吸去液体,将Transwell小室在盛有PBS的6 cm皿中刷洗三遍,不能碰到小室底部。将小室底部浸入结晶紫染色5~10 min(每孔1 mL)。纯净水洗小室,用棉签轻轻擦拭小室的上室,除去未与细胞结合的结晶紫;载玻片上加少量PBS,再将Transwell小室放上去,滴加PBS。在显微镜下取8个视野观察细胞并计数,进行比较。
1.2.6.4 平板克隆实验检测细胞的增殖能力
将处于对数生长期的细胞胰酶消化后,完全培养基(基础培养基+10%胎牛血清)重悬成细胞悬液,并计数;细胞接种于6孔板培养板中,各实验组接种800个细胞/孔;继续培养到14 d或绝大多数单个克隆中细胞数大于500为止,中途每隔3 d进行换液并观察细胞状态;克隆完成后,在显微镜下对细胞进行拍照,然后PBS洗涤1次,每孔加入1 mL 4%多聚甲醛固定30~60 min,PBS洗涤1次;每孔加入结晶紫染液1 mL,染色细胞10~20 min;PBS洗涤细胞数次,晾干,分别对整个6孔板及每个孔进行单独拍照。
1.2.6.5 RT-qPCR法检测基因表达
引物设计:下列引物基因由北京擎科生物科技股份有限公司(上海)进行合成。表2列出了引物的序列。
表 2 RT-qPCR引物序列Table 2. Sequence of primers for RT-qPCR引物名称 引物序列(5‘→3’) MAOA-F 5‘-ACAGCCTGACCGTGGAGAAG-3’ MAOA-R 5‘-GAACGGACGCTCCATTCGGA-3' AChE-F 5‘-GGGGTTGCCTGAGATTGGG-3' AChE-R 5‘-CGCAGGGACGACTTTCACA-3' CA2-F 5‘-GCCAAGTATGACCCTTCCCT-3’ CA2-R 5‘-TGTCCATCAAGTGAACCCCA-3' DPP4-F 5‘-CTGGGGTAAAGCCAGAATCA-3' DPP4-R 5‘-TGGTGGTGCACACCTGTAGT-'3 RNA提取:使用AGS细胞和MGC-803细胞铺于6孔板中,24 h后分别给予8、16 µmol/L的芹菜素继续培养24 h;将细胞从培养箱中取出,吸去培养基,使用PBS冲洗三次;每孔加入500 µL Trizol,充分吹打,使细胞充分裂解,然后转移到RNase-free 1.5 mL Ep管中,冰上静置10 min;每孔加入100 µL三氯甲烷,充分振荡15 s,室温静置15 min,12000 r/min,4 ℃离心15 min;离心后将上层水相转移到新的Ep管中,加等量的异丙醇并混匀,室温静置10 min,12000 r/min,4 ℃离心15 min;离心后弃上清,加500 µL预冷的75%乙醇,将沉淀振荡至悬浮,7500 r/min,4 ℃离心10 min;离心后弃上清,离心管倒置于面巾纸上,计时15 min,后将管口未干的液滴擦干;往Ep管中加入20 µL的DEPC水,充分吹打混合;最后使用核酸分析仪测定RNA浓度,吸取1 µL RNA进行测定,并分析纯度。
配制预混液:按表3顺序将表格中的成分依次加入到Ep管中,吹打均匀后离心。
表 3 预混液配方Table 3. Premix formula试剂名称 体积(µL) RNase-free dd H2O 至总体积20 2×One Step SYBR Green Mix 10 One Step SYBR Green Enzyme Mix 1 50×ROX Reference Dye 1 0.4 正向引物 0.4 反向引物 0.4 模版RNA 1 PCR反应:配制好后轻弹管底将溶液混合,6000 r/min短暂离心,离心结束后将96孔板置于RT-qPCR仪上。设置逆转录阶段温度为50 ℃,3 min;预变性阶段温度为95 ℃,30 s;PCR循环反应阶段为40个循环,温度设为95 ℃,10 s和60 ℃,30 s;溶解曲线阶段95 ℃,15 s;60 ℃,60 s;95 ℃,15 s;并缓慢加热至99 ℃。
1.3 数据处理
每组3个复孔重复3次,数据分析及绘图使用Image J和GraphPad Prism 9软件,结果使用平均值±标准差表示,其中,P<0.05表示有统计学意义。
2. 结果与分析
2.1 靶点的获取
以芹菜素作为研究对象,在ChEMBL数据库中搜索芹菜素的靶蛋白信息得到了共339个靶蛋白,从中筛选出Confidence 90%是“active、both”为有效靶蛋白,最后得到61个有效靶蛋白。
从GeneCards数据库中查询胃癌相关的基因,最后查到胃癌相关靶点总计15117个。
2.2 PPI网络的构建及成分-靶点网络图的构建
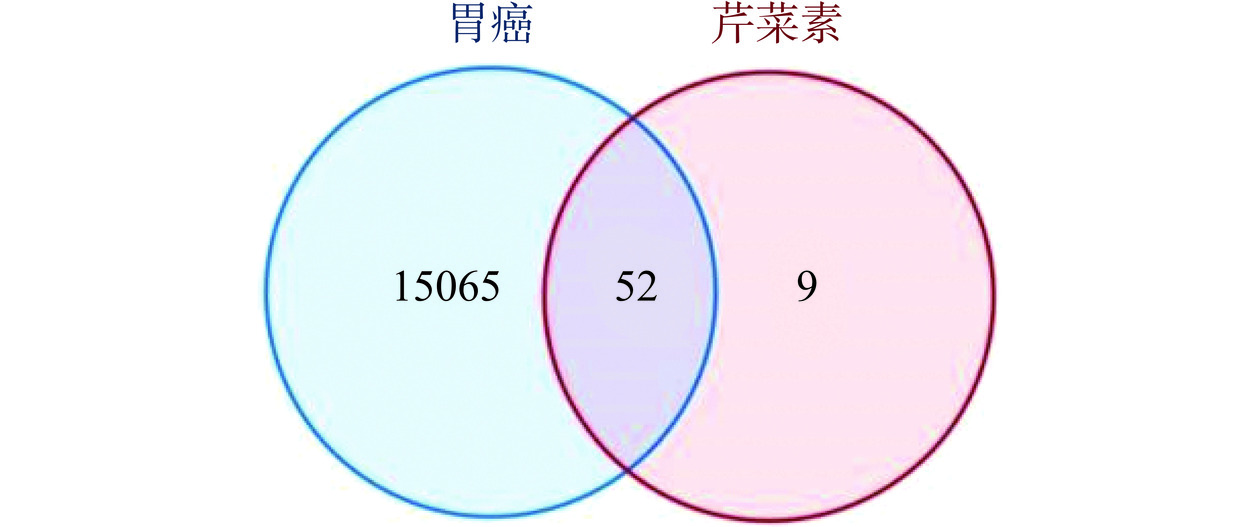
芹菜素61个有效靶点中交集得出与胃癌共同靶点52个,见图1,然后将共同靶点提交至STRING数据库,得到PPI网络,见图2。
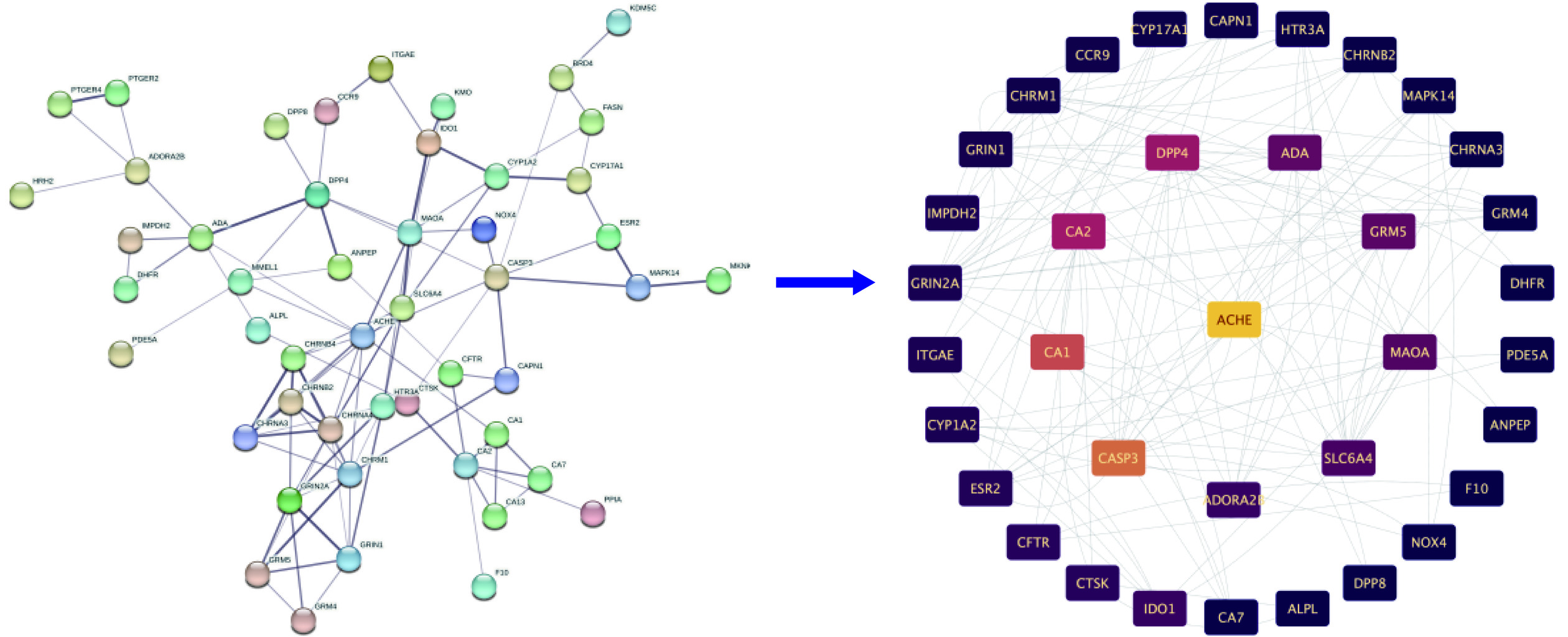
Cytoscape是一款可以将数据可视化的软件。将芹菜素与胃癌的相交靶点导入到Cytoscape 3.9.1软件中,构建成分-靶点网络图。筛选出“Betweenness”值大于200的靶点,最终得到芹菜素与胃癌相交靶点:AChE、CASP3、CA1、CA2、DPP4、ADA、GRM5、MAOA,共八个。成分靶点网络图见图2。
2.3 Go功能富集和KEGG通路富集分析
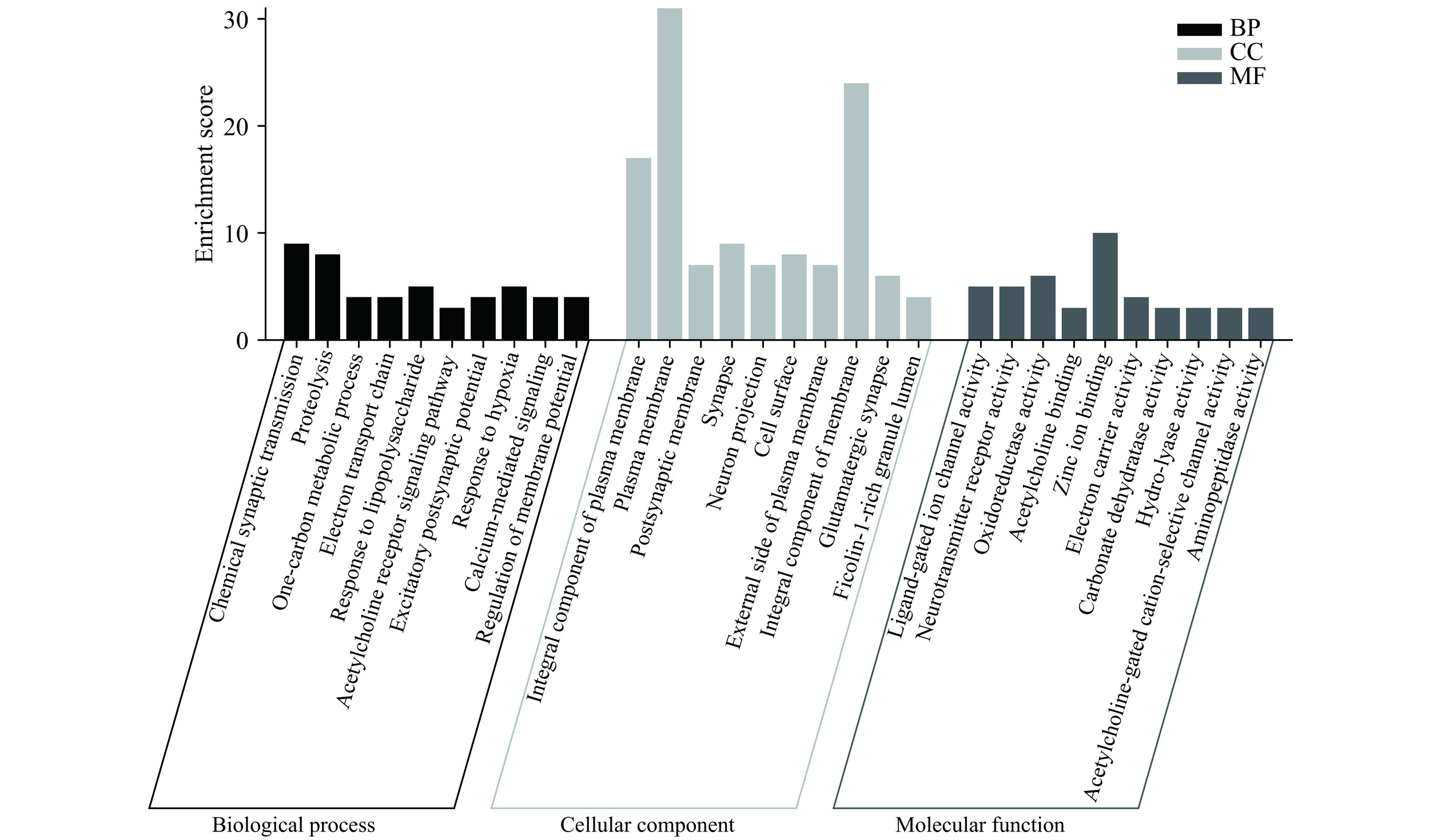
图3展示了芹菜素治疗胃癌的作用靶点GO功能富集结果。这些靶点参与信号传导、化学突触传递、蛋白水解、单碳代谢过程、质膜组成、质膜、突触后膜、配体门控离子通道活性、神经递质受体活性、氧化还原酶的活动等过程。
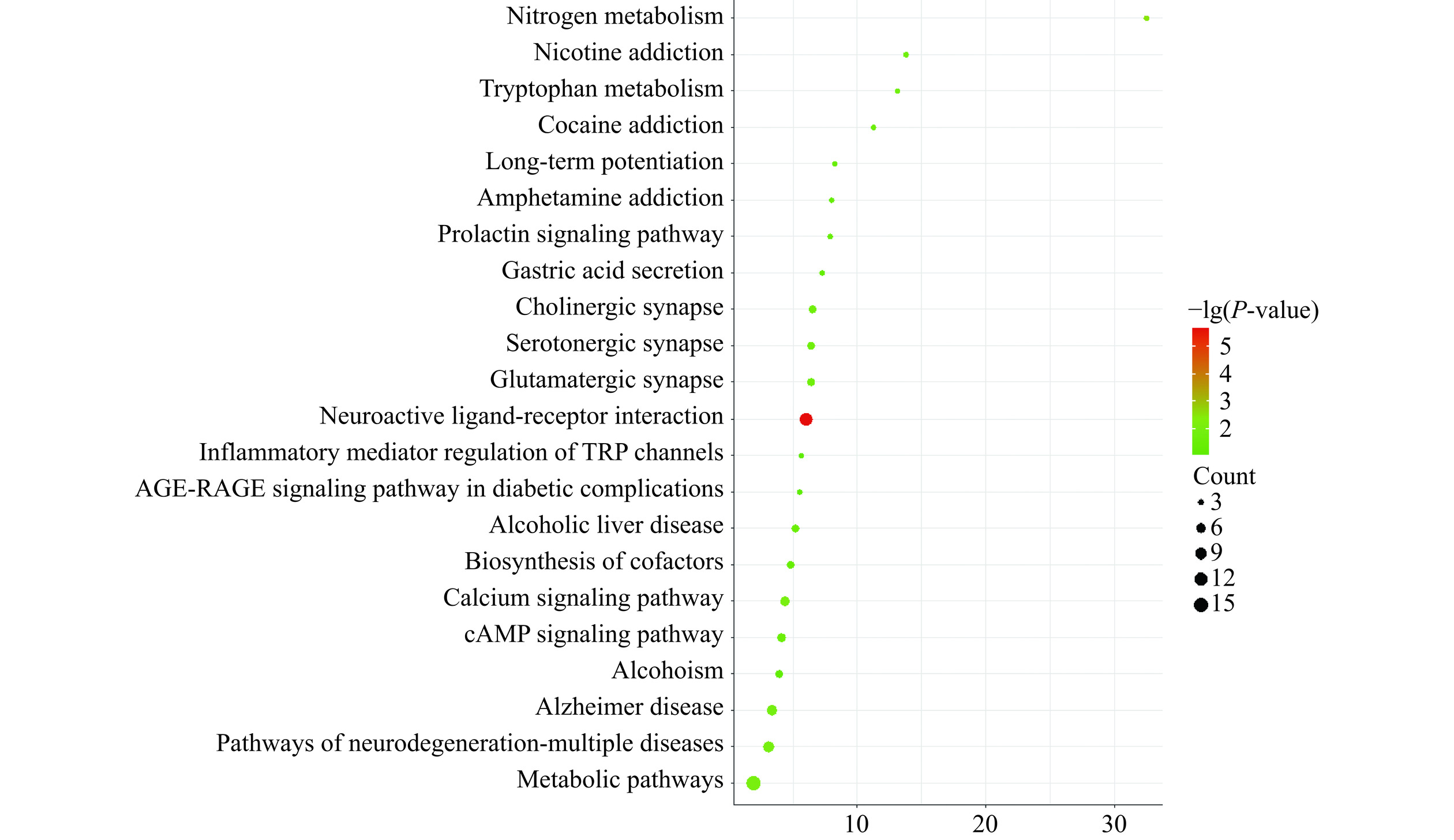
KEGG通路富集结果如图4所示,神经活性配体-受体相互作用、氮代谢、钙信号通路、代谢途径、神经退行性变的途径-多重疾病、阿尔茨海默病、色氨酸代谢、胆碱能突触、cAMP信号通路、酒精性肝病、胃酸分泌、酗酒等信号通路有靶点的富集。有相关研究发现,化学突触相关的通路P值越低,癌症存活率越小[19]。其中神经递质参与了多种恶性肿瘤的发生与发展[20],胆碱能信号抑制Wnt信号通路和干细胞的扩增,进而阻断胆碱能抑制胃癌的发生[21]。
2.4 分子对接
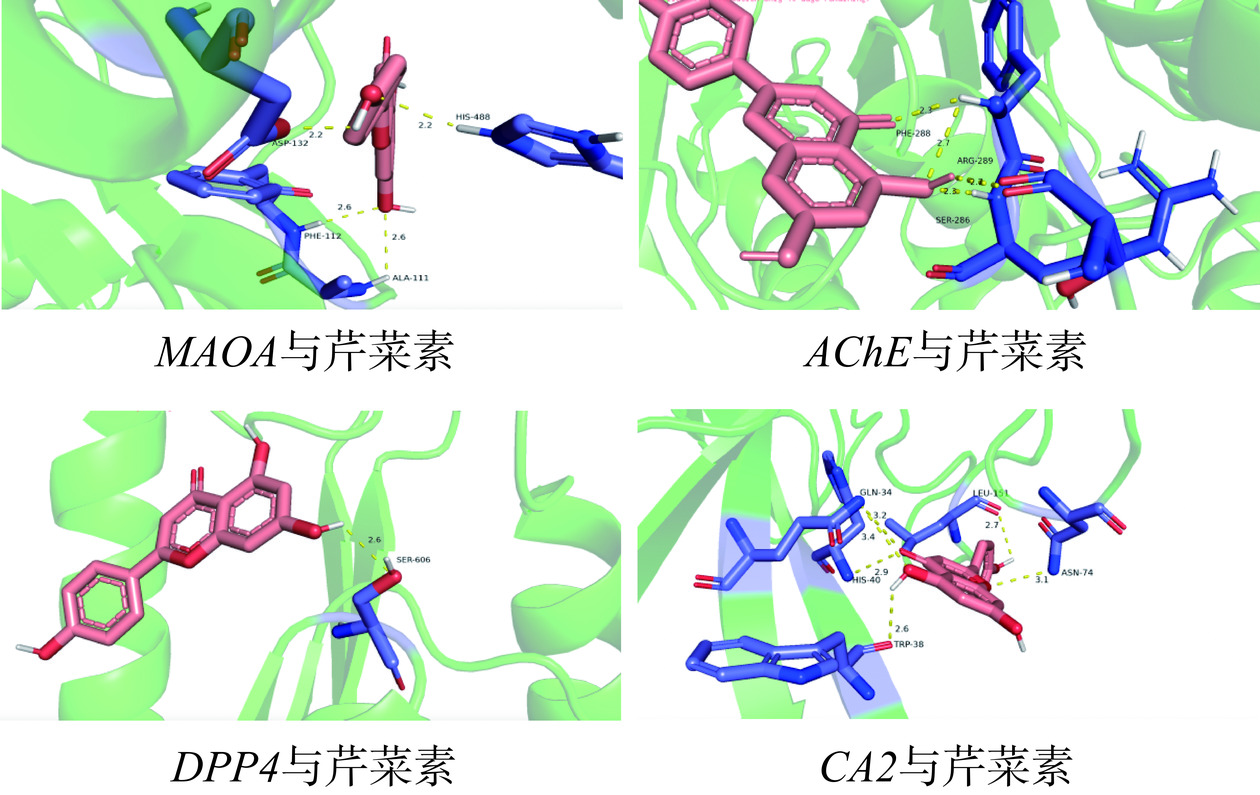
胃癌与芹菜素分子对接结果如表4所示,分子结合能绝对值的高低反映结合能力的强弱,绝对值排名为:AChE|−9.1|>MAOA|−8.8|>CA2|−7.4|>DPP4|−7.3|>CA1|−7.2|=GRM5|−7.2|>ADA|−6.8|>CASP3|−5.8|,可知与芹菜素结合紧密。表明芹菜素与胃癌中的以上靶点具有良好的结合能力,分子对接如图5所示。
表 4 胃癌靶点与芹菜素分子对接结果Table 4. Results of molecular docking between gastric cancer targets and apigenin靶点 结合能(kcal/mol) AChE −9.1 CASP3 −5.8 CA1 −7.2 CA2 −7.4 DPP4 −7.3 MAOA −8.8 ADA −6.8 GRM5 −7.2 2.5 体外实验
2.5.1 芹菜素对人胃癌细胞生存率的影响
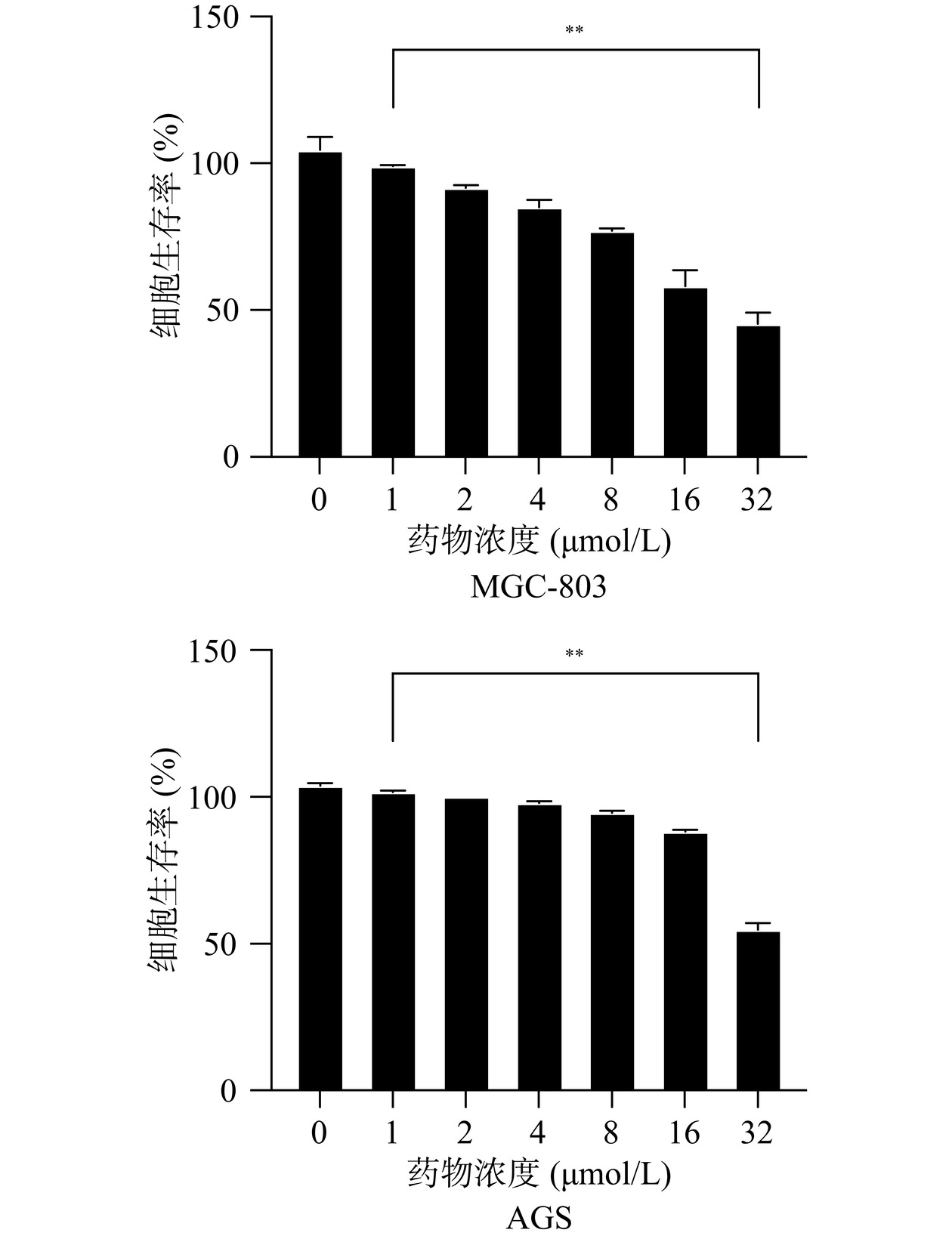
芹菜素常见于我们日常的食物里,主要以糖苷的形式存在。多项研究表明,芹菜素通过激活和抑制不同的细胞信号和分子来调节各种细胞过程,包括血管生成、凋亡、转移、自噬、细胞周期和免疫反应[22]。CCK-8检测细胞生存率在药物毒性检验、肿瘤细胞增殖等多个领域的研究中已经有所应用[23]。如图6所示,不同浓度的芹菜素处理的AGS细胞与MGC-803细胞的细胞活性呈剂量反比,芹菜素的浓度越高,细胞的生存率越低。通过各组之间比较得出,芹菜素可以降低胃癌细胞的存活率。由细胞生存率计算公式得出芹菜素对胃癌细胞系AGS的IC50为34.84 µmol/L,对MGC-803的IC50为24.86 µmol/L,MGC-803细胞的CCK-8结果中,与对照组相比8 µmol/L和16 µmol/L浓度的变化较明显,且与自身的IC50值的一半也较接近,浓度太高或太低可能会对细胞影响过大产生误差。因此选择8 µmol/L和16 µmol/L的给药浓度,继续做后续的实验。
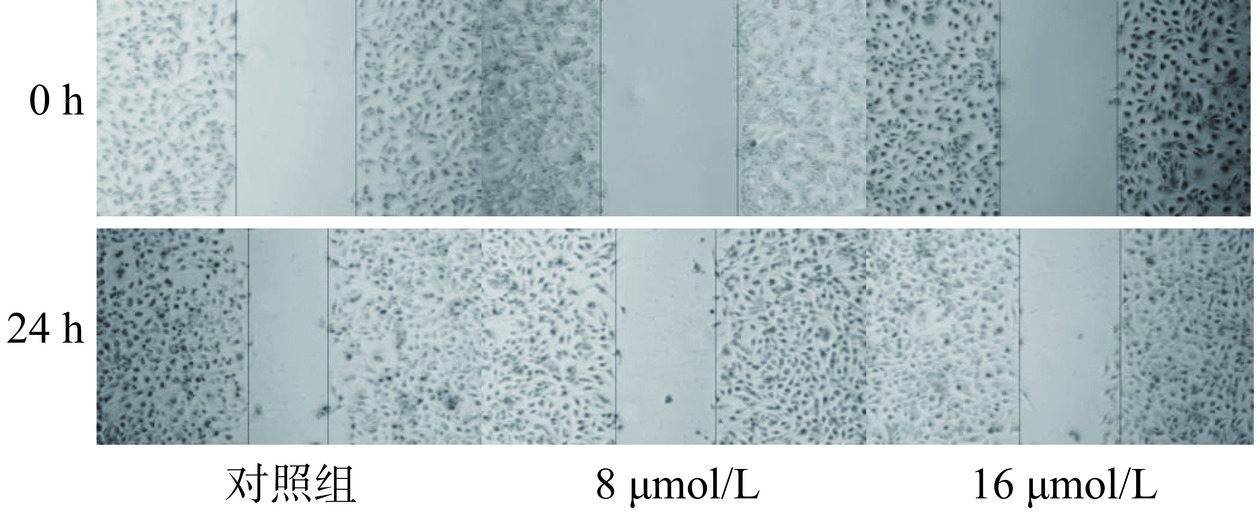
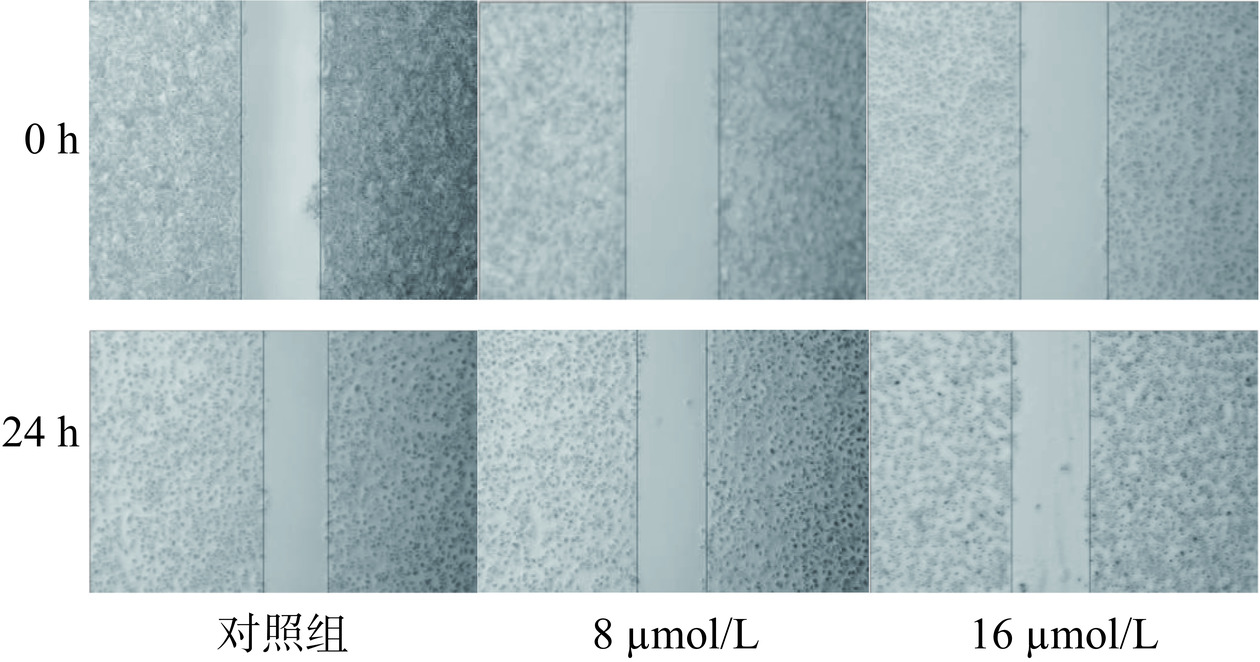
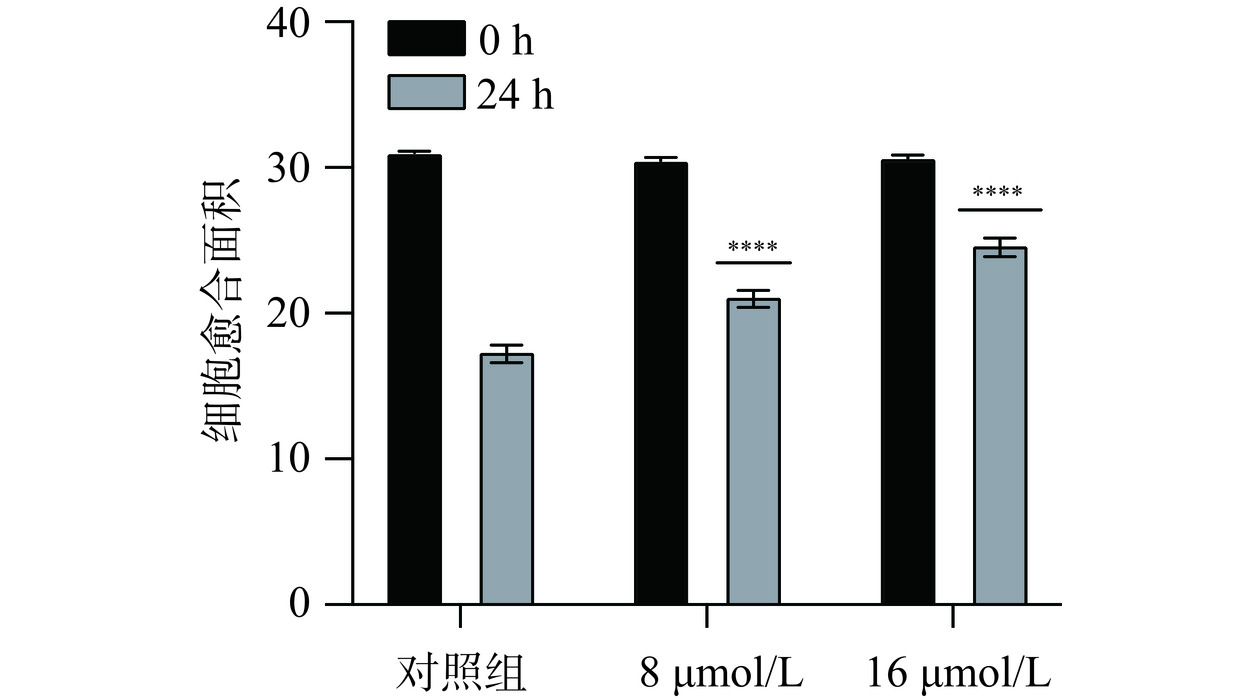
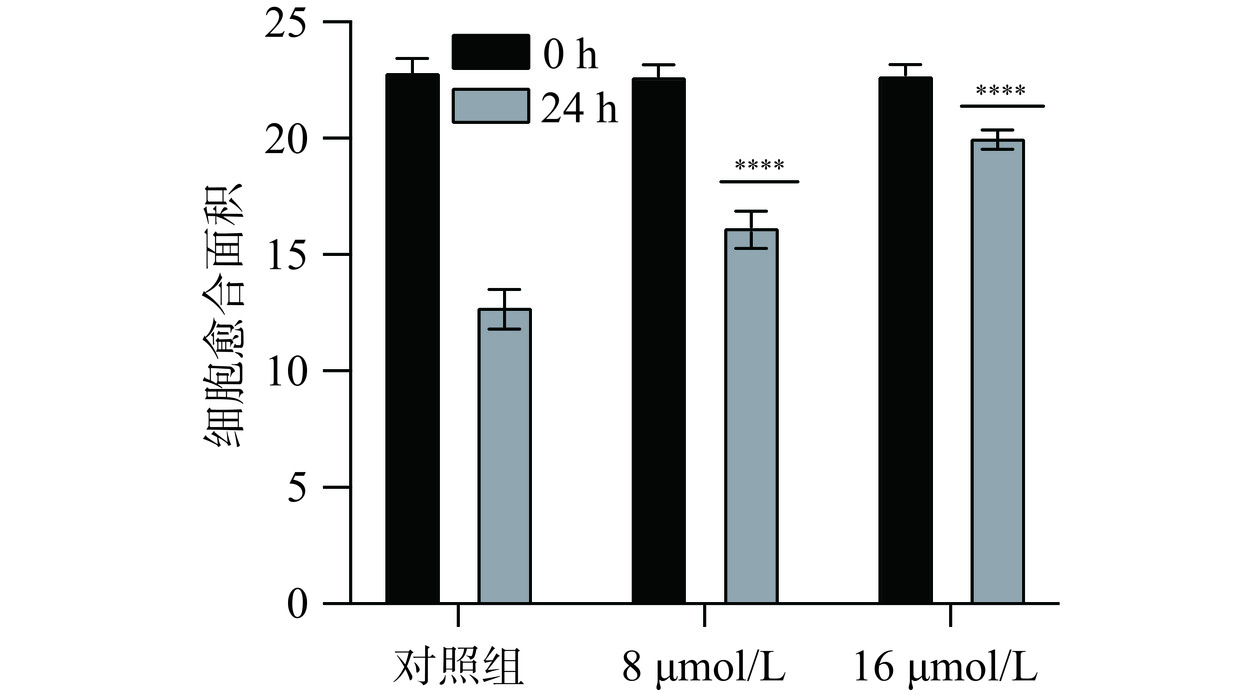
2.5.2 芹菜素对胃癌细胞愈合作用的影响
细胞划痕实验是通过在细胞单层上划痕并通过延时显微镜定期捕获图像,从而研究细胞迁移运动、修复能力和细胞间相互作用的实验室技术[24]。划痕实验结果如图7、图9所示,愈合结果柱状图如图8、图10所示。与对照组相比较,不同给药组的细胞划痕愈合率呈浓度依赖性,浓度越高,愈合率越低。通过该实验推测芹菜素可能通过干扰胃癌细胞的迁移和增殖相关机制,发挥其抑制愈合的效果。可能的作用机制包括调节细胞内的信号通路,如抑制与细胞增殖和迁移密切相关的PI3K/Akt/mTOR通路[25];或者诱导细胞凋亡,减少具有愈合能力的细胞数量。也有研究发现芹菜素可通过激活线粒体信号转导通路诱导人胃癌细胞BGC-823凋亡[26],并且可抑制胃癌细胞SGC-7901的增殖来诱导细胞凋亡[15]。
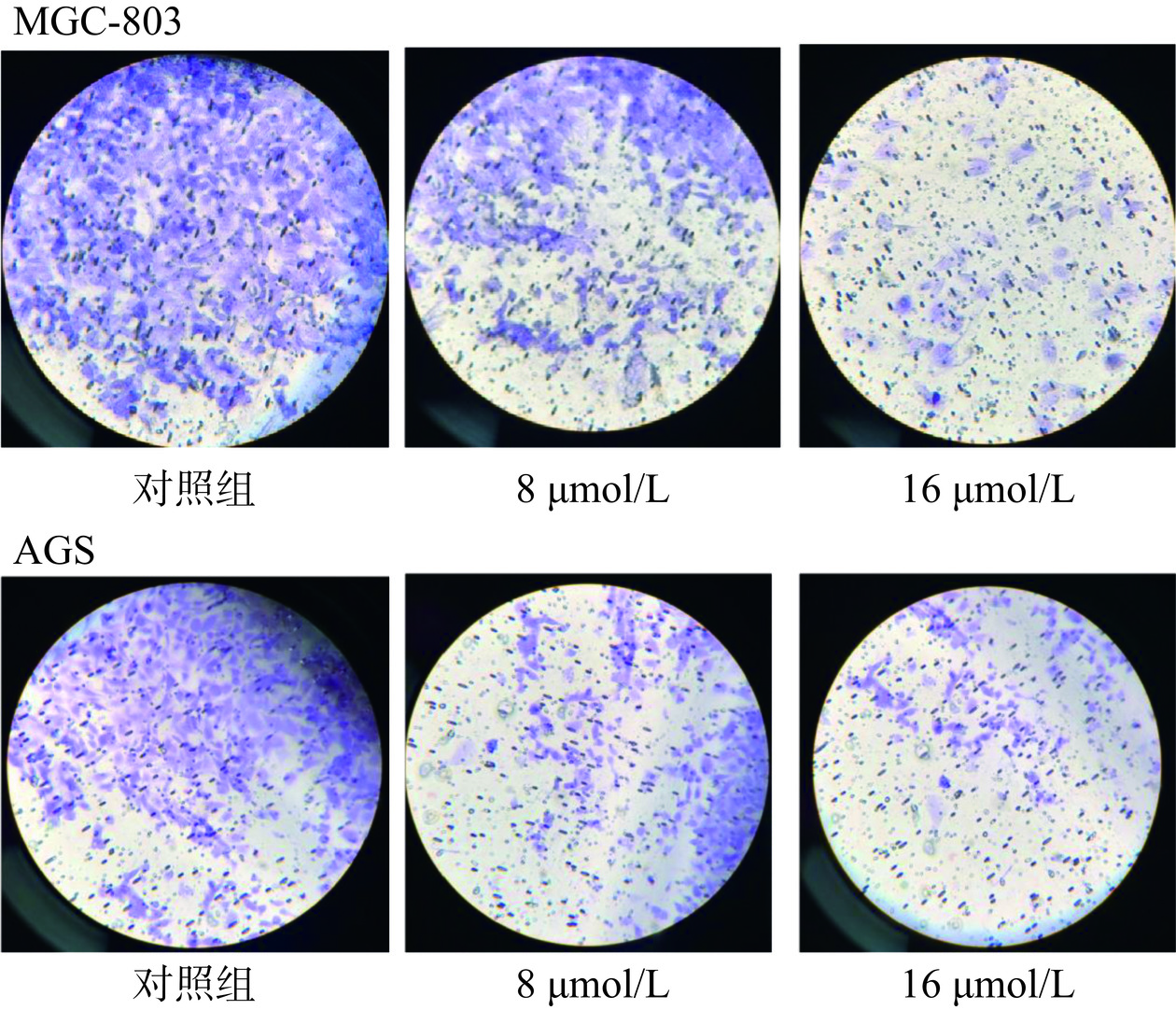
2.5.3 芹菜素对胃癌细胞侵袭的影响
细胞体外侵袭实验主要应用于肿瘤细胞侵袭和转移能力研究,该实验经常使用Transwell plate通过穿膜侵袭实验来检测肿瘤细胞的侵袭潜能[27]。图11所示,对照组及8、16 µmol/L芹菜素分别处理两种胃癌细胞24 h后,细胞侵袭数量发生明显变化。MGC-803对照组细胞侵袭数量832个,8 µmol/L芹菜素组389个,16 µmol/L芹菜素组减少至82个;AGS对照组细胞侵袭数量632个,8 µmol/L芹菜素组276个,16 µmol/L芹菜素组65个。上述结果表明芹菜素处理对胃癌细胞的侵袭能力产生明显影响,并存在一定的浓度依赖性。
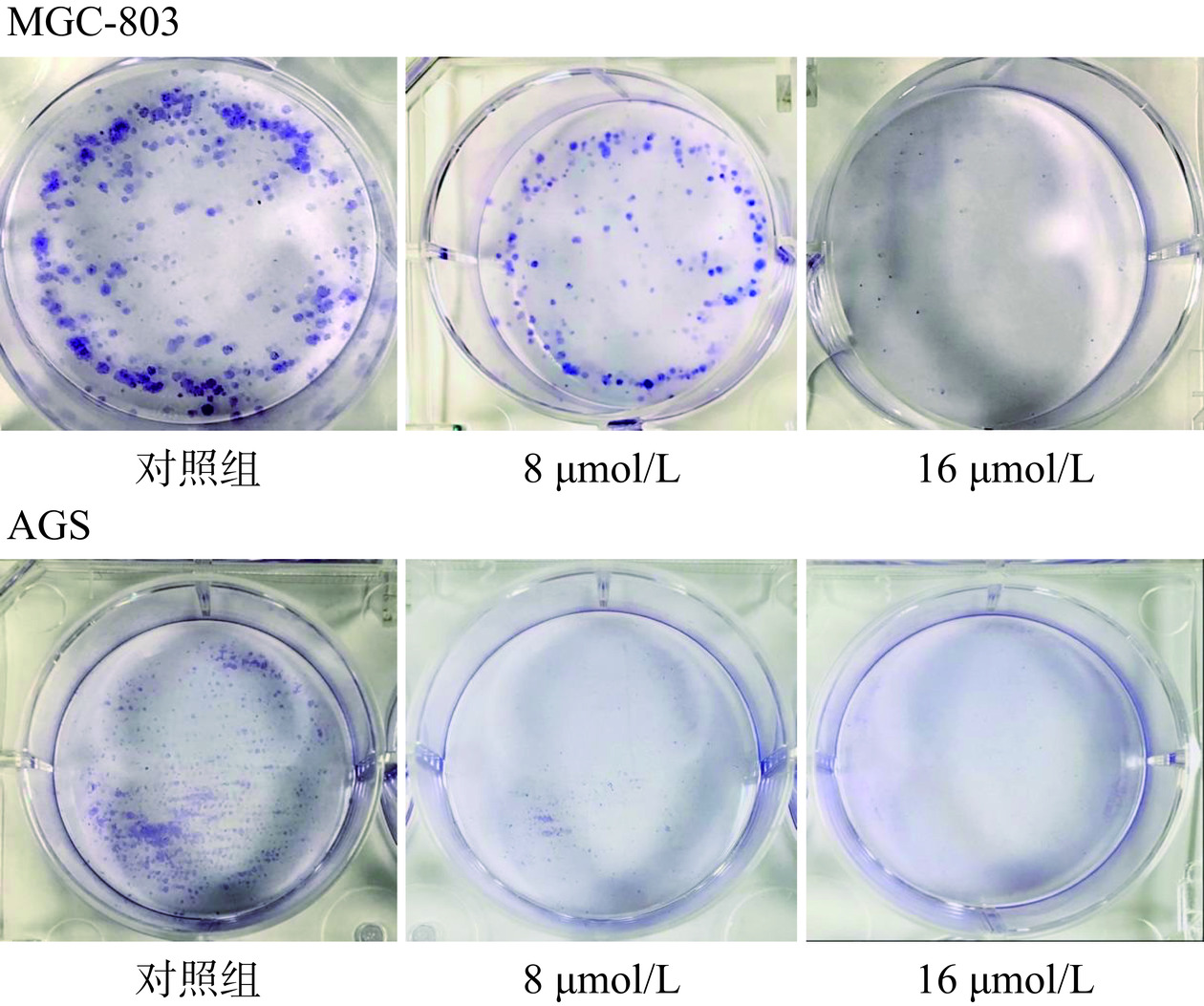
2.5.4 细胞克隆形成实验
当单个细胞在体外增殖六代以上,其后代所组成的细胞群体,成为集落或群体[28]。实验结果如图12所示,与对照组相比,可以看到不同浓度的芹菜素组中的克隆细胞数明显减少直至几乎无,并且每个克隆细胞群的体积也随着芹菜素的浓度增加而减少。
2.5.5 RT-qPCR结果
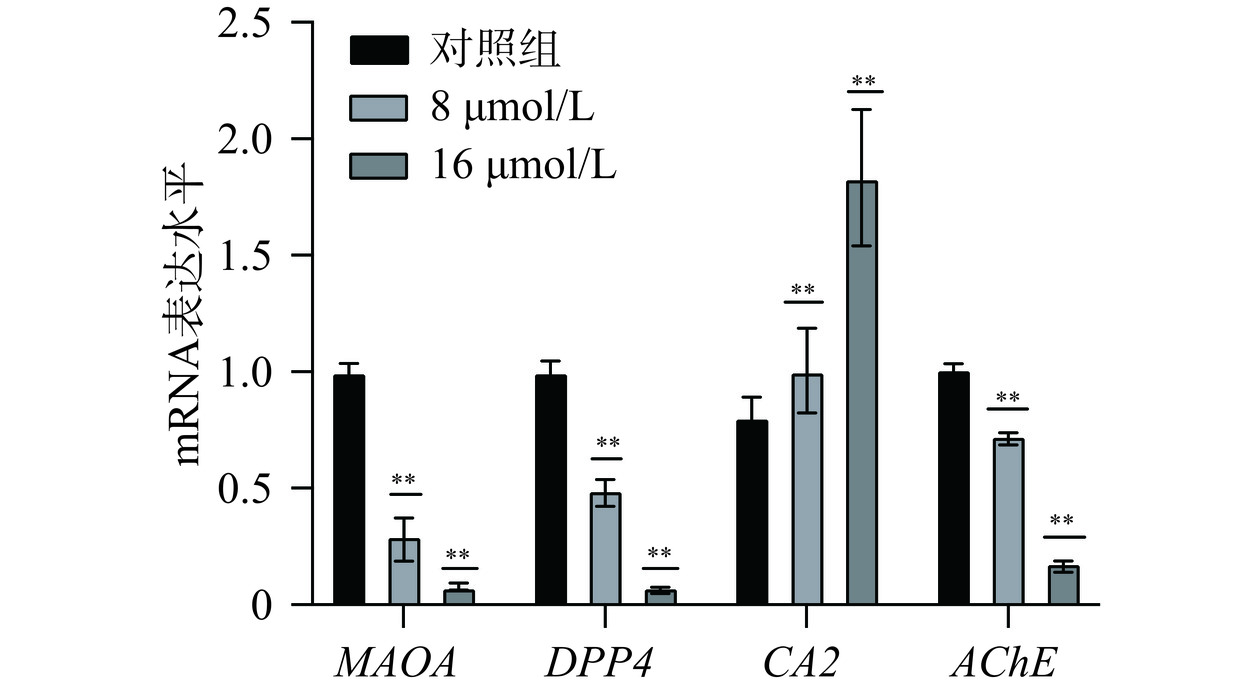
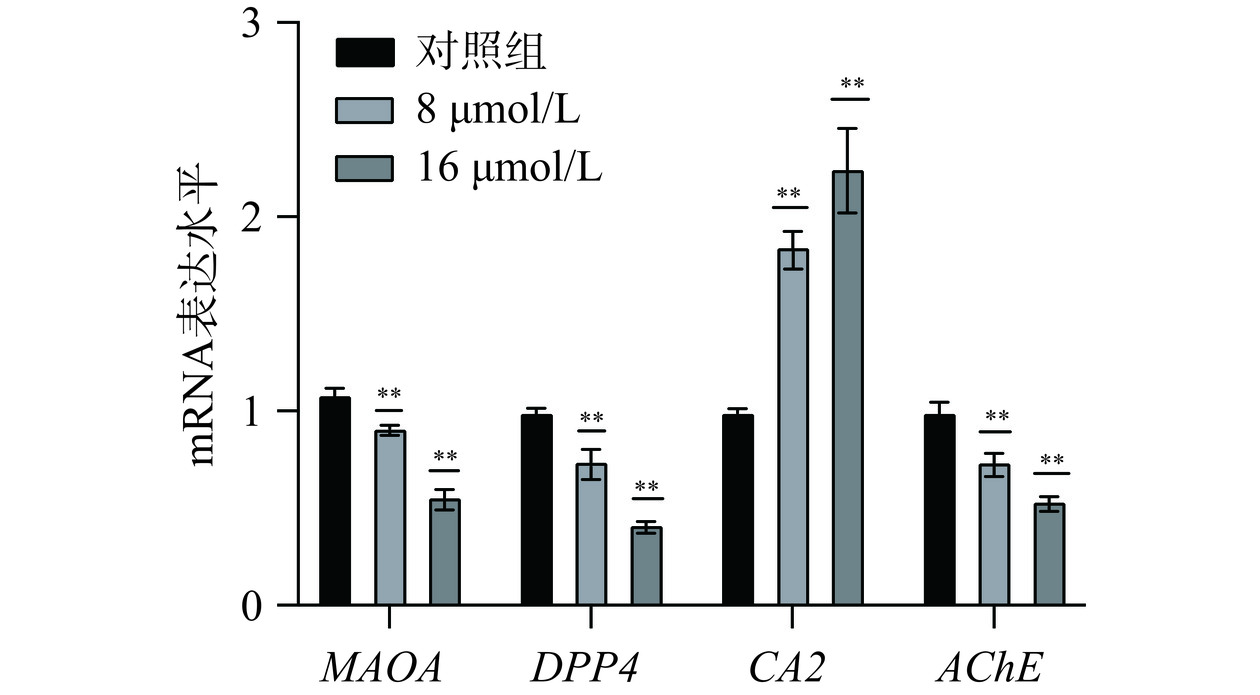
为了进一步验证芹菜素对关键基因的调控作用,采用RT-qPCR分析人胃癌细胞系MGC-803和AGS细胞系中MAOA、DPP4、CA2、AChE的mRNA表达水平。RT-qPCR结果如图13~图14所示,与对照组相比,使用芹菜素处理后,基因CA2的表达水平显著上调(P<0.01)。基因MAOA、DPP4、AChE表达水平受到显著抑制,表达降低(P<0.01)。MAOA(单胺氧化酶A)可通过氧化脱氨作用降解胺类神经递质[29],DPP4(二肽基肽酶4)是广泛表达的跨膜糖蛋白[30],可作为潜在的泛癌生物标志物[31]。AChE(乙酰胆碱酯酶)也在生物神经传导中起着关键性的作用[32]。AChE是一种分泌型羧酸酯酶,能够降解神经递质乙酰胆碱成为胆碱和乙酸[33]。AChE可以使干细胞分化[34]、突触发生[35]、促进细胞生长、在细胞凋亡的过程中起着重要作用[36]。CA(碳酸酐酶)是含有可催化二氧化碳可逆水合作用的含锌金属酶[37]。推测芹菜素可能是通过促进CA2的表达水平,抑制MAOA、DPP4、AChE的表达水平来达到治疗胃癌的作用,且以上四个靶点参与免疫调节,通过物质代谢、炎症反应等相关通路调控肿瘤的发生发展。
3. 结论
本文采用网络药理学筛选出芹菜素共61个有效靶点,与胃癌交叉靶点有52个。芹菜素靶点与抗胃癌靶点PPI网络分析为深入研究芹菜素对胃癌细胞的作用机制提供了重要线索,也为开发新的抗胃癌治疗策略提供了潜在的方向;GO富集分析说明芹菜素抗胃癌的作用与生物学方面有关;KEGG通路分析表明抗癌机制与神经递质、胆碱能、物质代谢和癌症的发展相关。而芹菜素具体是通过哪一类信号通路抑制胃癌还需进一步的深刻研究。
本文通过网络药理学筛选、分子对接技术,预测了芹菜素治疗胃癌的可能靶点。通过神经递质、胆碱能、物质代谢等相关通路来抗胃癌,体外细胞实验验证了芹菜素能够有效地抑制胃癌细胞MGC-803和AGS的增殖、迁移、形成群落能力与侵袭,后续可在本研究的基础上,增加动物实验进行验证,进一步明确芹菜素抗胃癌的作用机制。
-
表 1 网络药理学分析的参照数据库及网址
Table 1 Database and website for network pharmacology analysis
数据库 名称 网址 ChEMBL 药物化学数据库 https://www.ebi.ac.uk/chembl/ GeneCards 人类基因综合数据库 https://www.genecards.org UniProt 蛋白质信息数据库 https://www.uniprot.org jvenn 在线韦恩图 https://jvenn.toulouse.inrae.fr/ STRING 蛋白互作数据库 https://cn.string-db.org David 注释、可视化和综合发现数据库 https://david.ncifcrf.gov PDB 国际蛋白质结构数据库 https://www.rcsb.org Bioinformatics 微生信数据平台 http://www.bioinformatics.com.cn 表 2 RT-qPCR引物序列
Table 2 Sequence of primers for RT-qPCR
引物名称 引物序列(5‘→3’) MAOA-F 5‘-ACAGCCTGACCGTGGAGAAG-3’ MAOA-R 5‘-GAACGGACGCTCCATTCGGA-3' AChE-F 5‘-GGGGTTGCCTGAGATTGGG-3' AChE-R 5‘-CGCAGGGACGACTTTCACA-3' CA2-F 5‘-GCCAAGTATGACCCTTCCCT-3’ CA2-R 5‘-TGTCCATCAAGTGAACCCCA-3' DPP4-F 5‘-CTGGGGTAAAGCCAGAATCA-3' DPP4-R 5‘-TGGTGGTGCACACCTGTAGT-'3 表 3 预混液配方
Table 3 Premix formula
试剂名称 体积(µL) RNase-free dd H2O 至总体积20 2×One Step SYBR Green Mix 10 One Step SYBR Green Enzyme Mix 1 50×ROX Reference Dye 1 0.4 正向引物 0.4 反向引物 0.4 模版RNA 1 表 4 胃癌靶点与芹菜素分子对接结果
Table 4 Results of molecular docking between gastric cancer targets and apigenin
靶点 结合能(kcal/mol) AChE −9.1 CASP3 −5.8 CA1 −7.2 CA2 −7.4 DPP4 −7.3 MAOA −8.8 ADA −6.8 GRM5 −7.2 -
[1] 刘德华. 胃癌发生发展相关胃肠道菌群以及SFRPs基因的生物信息研究[D]. 合肥:中国科学技术大学, 2022. [LIU Dehua, Bioinformatics research on the gastrointestinal microbiota and SFRPs genes related to the occurrence and development of gastric cancer[D]. Hefei:University of Science and Technology of China, 2022.] LIU Dehua, Bioinformatics research on the gastrointestinal microbiota and SFRPs genes related to the occurrence and development of gastric cancer[D]. Hefei: University of Science and Technology of China, 2022.
[2] MUHAMMAD I, TANWEER A G, MUHAMMAD A, et al. Apigenin as an anticancer agent[J]. Phytother Res,2020,34(8):1812−1828. doi: 10.1002/ptr.6647
[3] 计荣盛, 王梓灵, 吴婷, 等. 芹菜素与氧化苦参碱联合用药抑制非小细胞肺癌的作用及其机制研究[J]. 中国中药杂志,2023,48(3):752−761. [JI Rongsheng, WANG Ziling, WU Ting, et al. Effect of apigenin in combination with oxymatrine on non-small cell lung cancer and mechanism[J]. China Journal of Chinese Materia Medica,2023,48(3):752−761.] JI Rongsheng, WANG Ziling, WU Ting, et al. Effect of apigenin in combination with oxymatrine on non-small cell lung cancer and mechanism[J]. China Journal of Chinese Materia Medica, 2023, 48(3): 752−761.
[4] FARIEHA H, NAZISH J, KHALIL UR-R, et al. Identification of hypotensive biofunctional compounds of Coriandrum sativum and evaluation of their angiotensin-converting enzyme (ACE) inhibition potential[J]. Oxid Med Cell Longev,2018:4643736.
[5] SARA P, MARIKA P, ANTONELLA R, et al. Chemical composition and antioxidant activity of thyme, hemp and coriander extracts:A comparison study of maceration, soxhlet, UAE and RSLDE techniques[J]. Foods,2020,9(9):9091221.
[6] 杨文志, 杜跃中, 高宇, 等. 蒲公英总黄酮的研究进展[J]. 人参研究,2017,15(1):52−55. [YANG Wenzhi, DU Yuezhong, GAO Yu, et al. The research progress of dandelion flavonoids[J]. Ginseng Research,2017,15(1):52−55.] YANG Wenzhi, DU Yuezhong, GAO Yu, et al. The research progress of dandelion flavonoids[J]. Ginseng Research, 2017, 15(1): 52−55.
[7] 冷新, 倪健, 游龙泰, 等. HPLC法测定德国洋甘菊中槲皮素、木犀草素及芹菜素含量[J]. 国际中医中药杂志,2018,40(7):649−652. [LENG Xin, NI Jian, YOU Longtai, et al. Determination of quercetin, luteolin and apigenin in German chamomile by HPLC[J]. International Journal of Traditional Chinese Medicine,2018,40(7):649−652.] doi: 10.3760/cma.j.issn.1673-4246.2018.07.016 LENG Xin, NI Jian, YOU Longtai, et al. Determination of quercetin, luteolin and apigenin in German chamomile by HPLC[J]. International Journal of Traditional Chinese Medicine, 2018, 40(7): 649−652. doi: 10.3760/cma.j.issn.1673-4246.2018.07.016
[8] 陈红影, 苗培培, 郭嫦娥, 等. 芫花主要黄酮成分对肝微粒体UGTs及UGT1A1活性的体外抑制作用[J]. 中国现代应用药学,2017,34(3):305−310. [CHEN Hongying, MIAO Peipei, GUO Change, et al. In vitro inhibitory effect of main flavonoids ingredients of genkwa flos on liver microsomal UGTs and UGT1A1 activities[J]. Chinese Journal of Modern Applied Pharmacy,2017,34(3):305−310.] CHEN Hongying, MIAO Peipei, GUO Change, et al. In vitro inhibitory effect of main flavonoids ingredients of genkwa flos on liver microsomal UGTs and UGT1A1 activities[J]. Chinese Journal of Modern Applied Pharmacy, 2017, 34(3): 305−310.
[9] GEETHI P, D NEDRA K, VIDURANGA Y W. Antidiabetic properties, bioactive constituents, and other therapeutic effects of Scoparia dulcis[J]. Evid Based Complement Alternat Med,2016,2016(8):8243215.
[10] ZENG Y Z, TIAN G, YAN H, et al. Apigenin ameliorates hypertension-induced cardiac hypertrophy and down-regulates cardiac hypoxia inducible factor-lα in rats[J]. Food Funct,2016,7(4):1992−1998. doi: 10.1039/C5FO01464F
[11] PENGFEI C, XIANHAO H, WENQING L, et al. Apigenin exhibits anti-inflammatory effects in LPS-stimulated BV2 microglia through activating GSK3β/Nrf2 signaling pathway[J]. Immunopharmacol Immunotoxicol,2020,42(1):9−16. doi: 10.1080/08923973.2019.1688345
[12] PIYUSH K, DEEP S, MAMTA T, et al. Functionality of apigenin as a potent antioxidant with emphasis on bioavailability, metabolism, action mechanism and in vitro and in vivo studies:A review[J]. Food Biochem,2022,46(4):e13950.
[13] GITIRANA DE SANTANA J D, SANTOS-MAYORGA O A, FLORENCIO J R, et al. Vernonia polyanthes Less. (Asteraceae Bercht. & Presl), a natural source of bioactive compounds with antibiotic effect against multidrug-resistant Staphylococcus aureus[J]. Antibiotics (Basel),2023,12(3):622. doi: 10.3390/antibiotics12030622
[14] 林沈国, 毛怡雯, 赵亚新. 芹菜素抗乳腺癌作用及机制的研究进展[J]. 浙江医学,2023,45(2):213−219. [LIN Shenguo, MAO Yiwen, ZHAO Yaxin. Research progress of apigenin anti-breast cancer effect and mechanism[J]. Zhejiang Medical Journal,2023,45(2):213−219.] doi: 10.12056/j.issn.1006-2785.2023.45.2.2022-2590 LIN Shenguo, MAO Yiwen, ZHAO Yaxin. Research progress of apigenin anti-breast cancer effect and mechanism[J]. Zhejiang Medical Journal, 2023, 45(2): 213−219. doi: 10.12056/j.issn.1006-2785.2023.45.2.2022-2590
[15] 崔昭阳, 李素娜, 姜旭倩, 等. 芹菜素激活TRAIL死亡受体参与诱导胃癌SGC-7901细胞凋亡的实验研究[J]. 癌变·畸变·突变,2021,33(1):32−36. [CUI Zhaoyang, LI Suna, JIANG Xuqian, et al. Activation of the TRAIL death receptor by apigenin for induction of apoptosis in the gastric cancer SGC-7901 cells[J]. Carcinogenesis, Teratogenesis & Mutagenesis,2021,33(1):32−36.] CUI Zhaoyang, LI Suna, JIANG Xuqian, et al. Activation of the TRAIL death receptor by apigenin for induction of apoptosis in the gastric cancer SGC-7901 cells[J]. Carcinogenesis, Teratogenesis & Mutagenesis, 2021, 33(1): 32−36.
[16] 崔迎红, 陈阿, 许畅, 等. 芹菜素上调SHP-1蛋白表达抑制肝癌MHCC97H细胞STAT3磷酸化及球形成[J]. 湖南师范大学学报(医学版),2018,15(5):1−4. [CUI Yinghong, CHEN E, XU Chang, et al. Inhibition of spheroid formation and STAT3 phosphorylation through up-regulating expression of SHP-1 by apigenin in human liver cancer MHCC97H cells[J]. Journal of Hunan Normal University (Medical science),2018,15(5):1−4.] doi: 10.3969/j.issn.1673-016X.2018.05.001 CUI Yinghong, CHEN E, XU Chang, et al. Inhibition of spheroid formation and STAT3 phosphorylation through up-regulating expression of SHP-1 by apigenin in human liver cancer MHCC97H cells[J]. Journal of Hunan Normal University (Medical science), 2018, 15(5): 1−4. doi: 10.3969/j.issn.1673-016X.2018.05.001
[17] 谢孟君. 基于网络药理学和代谢组学技术对补肾活血方防治DR的作用机制及配伍机制研究[D]. 成都:成都中医药大学, 2021. [XIE Mengjun. Study on the action mechanism and compatibility mechanism of bushen huoxue prescription in the prevention and treatment of DR based on network pharmacology and metabonomics[D]. Chengdu:Chengdu University of Traditional Chinese Medicine, 2021.] XIE Mengjun. Study on the action mechanism and compatibility mechanism of bushen huoxue prescription in the prevention and treatment of DR based on network pharmacology and metabonomics[D]. Chengdu: Chengdu University of Traditional Chinese Medicine, 2021.
[18] ZHAO L, ZHANG H, LI N, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula[J]. J Ethnopharmacol,2023,309(12):116306.
[19] 金其煌. 乙酰胆碱酯酶在细胞凋亡中的作用及G418诱导细胞凋亡的分子机制[D]. 上海:中国科学院研究生院(上海生命科学研究院), 2004. [JIN Qihuang. The role of acetylcholinesterase in apoptosis[D]. Shanghai:Shanghai Institutes for Biological Sciences, 2004.] JIN Qihuang. The role of acetylcholinesterase in apoptosis[D]. Shanghai: Shanghai Institutes for Biological Sciences, 2004.
[20] 王飞龙. 神经系统在癌症中的功能研究及生物标志物识别[D]. 长春:吉林大学, 2022. [WANG Feilong. Function research and biomarker identification of nervous system in cancer[D]. Changchun:Jilin University, 2022.] WANG Feilong. Function research and biomarker identification of nervous system in cancer[D]. Changchun: Jilin University, 2022.
[21] 周辉. 神经相关因子在结直肠癌患者中的表达及迷走神经对免疫微环境的影响[D]. 石家庄:河北医科大学, 2021. [ZHOU Hui. Expression of eurotransmitters in colorectal cancer and effect of vagus nerve on immune microenvironment[D]. Shijiazhuang:Hebei Medical University, 2021.] ZHOU Hui. Expression of eurotransmitters in colorectal cancer and effect of vagus nerve on immune microenvironment[D]. Shijiazhuang: Hebei Medical University, 2021.
[22] 黄杰波, 贾海英, 张涛. CAII与恶性肿瘤的相关研究[J]. 世界最新医学信息文摘,2019,19(43):128−130. [HUANG Jiebo, JIA Haiying, ZHANG Tao. Correlation between CAII and malignant tumors[J]. World Latest Medicine Information,2019,19(43):128−130.] HUANG Jiebo, JIA Haiying, ZHANG Tao. Correlation between CAII and malignant tumors[J]. World Latest Medicine Information, 2019, 19(43): 128−130.
[23] 金雅晴. 基于网络药理学的抗流感病毒药物研究[D]. 北京:军事科学院, 2022. [[JIN Yaqing. Network-based pharmacology of anti-influenza virus drugs:The case of moslae herba[D]. Beijing:Military science, 2022.] [JIN Yaqing. Network-based pharmacology of anti-influenza virus drugs: The case of moslae herba[D]. Beijing: Military science, 2022.
[24] 关玮, 杨树国, 李健, 等. CCK-8法在阴道毛滴虫体外增殖检测中的应用研究[J]. 湖北医药学院学报,2021,40(6):591−596. [GUAN Wei, YANG Shuguo, LI Jian, et al. The application of CCK-8 method in the detection of trichomonas proliferation in vitro[J]. Journal of Hubei University of Medicine,2021,40(6):591−596.] GUAN Wei, YANG Shuguo, LI Jian, et al. The application of CCK-8 method in the detection of trichomonas proliferation in vitro[J]. Journal of Hubei University of Medicine, 2021, 40(6): 591−596.
[25] 孙悦, 余洋, 吴坤. 芹菜素诱导胃癌SGC-7901细胞自噬及其对凋亡的影响[J]. 癌变·畸变·突变,2017,29(4):289−294, 299. [SUN Yue, YU Yang, WU Kun. Effect of apeginin-induced autophagy on apoptosis in human gastric cancer cells SGC-7901[J]. Carcinogenesis, Teratogenesis & Mutagenesis,2017,29(4):289−294, 299.] SUN Yue, YU Yang, WU Kun. Effect of apeginin-induced autophagy on apoptosis in human gastric cancer cells SGC-7901[J]. Carcinogenesis, Teratogenesis & Mutagenesis, 2017, 29(4): 289−294, 299.
[26] 黄暨生. 木犀草素、芹菜素及槲皮素对黑色素瘤自杀基因治疗的增效研究[D]. 广州:广州中医药大学, 2016. [HUANG Jisheng. Study on synergism of luteolin, apigenin and quercetin on suicide gene therapy of melanoma[D]. Guangzhou:Guangzhou University of Chinese Medicine, 2016.] HUANG Jisheng. Study on synergism of luteolin, apigenin and quercetin on suicide gene therapy of melanoma[D]. Guangzhou: Guangzhou University of Chinese Medicine, 2016.
[27] DENNIS K, WOLFGANG W, ULRIKE S S. Real-time cell migration monitoring to analyze drug synergism in the scratch assay using the incucyte system[J]. Methods Mol Biol,2021,2294(3):133−142.
[28] 蒋海鹰, 颜亚晖, 罗庚求, 等. 细胞体外侵袭实验中Transwell plate染色技术改良[J]. 中国组织化学与细胞化学杂志,2011,96(6):644−645. [JIANG Haiying, YAN Yahui, LUO Gengqiu, et al. Improved Transwell plate staining technique in cell invasion in vitro[J]. Chinese Journal of Histochemistry and Cytochemistry,2011,96(6):644−645.] doi: 10.3870/zgzzhx.2011.06.022 JIANG Haiying, YAN Yahui, LUO Gengqiu, et al. Improved Transwell plate staining technique in cell invasion in vitro[J]. Chinese Journal of Histochemistry and Cytochemistry, 2011, 96(6): 644−645. doi: 10.3870/zgzzhx.2011.06.022
[29] 刘震. SHP2抑制剂Batoprotafib治疗耐药胃肠间质瘤(GIST)的机制研究及GIST综合治疗分析[D]. 北京:北京协和医学院, 2023. [LIU Zhen. SHP2 allosteric inhibitor Batoprotafib inhibits prolifeation of imatinib-resistant gastrointestinal stromal tumor (GIST) and the comprehensive analyses of treatment strategies for GIST[D]. Beijing:Peking Union Medical College, 2023.] LIU Zhen. SHP2 allosteric inhibitor Batoprotafib inhibits prolifeation of imatinib-resistant gastrointestinal stromal tumor (GIST) and the comprehensive analyses of treatment strategies for GIST[D]. Beijing: Peking Union Medical College, 2023.
[30] 黄炳御, 唐旭东. MAOA与肿瘤关系的研究进展[J]. 生命科学,2018,30(5):545−550. [HUANG Bingyu, TANG Xudong. Progress in the relationship between MAOA and tumor[J]. Chinese Bulletin of Life Sciences,2018,30(5):545−550.] HUANG Bingyu, TANG Xudong. Progress in the relationship between MAOA and tumor[J]. Chinese Bulletin of Life Sciences, 2018, 30(5): 545−550.
[31] 谢云亮, 赵云娟, 李万根. 二肽基肽酶4相关信号通路研究新进展[J]. 广东医学,2018,39(12):1901−1905. [XIE Yunliang, ZHAO Yunjuan, LI Wangen. New advances in the study of dipeptidyl peptidase 4 related signaling pathways[J]. Guangdong Medical Journal,2018,39(12):1901−1905.] doi: 10.3969/j.issn.1001-9448.2018.12.037 XIE Yunliang, ZHAO Yunjuan, LI Wangen. New advances in the study of dipeptidyl peptidase 4 related signaling pathways[J]. Guangdong Medical Journal, 2018, 39(12): 1901−1905. doi: 10.3969/j.issn.1001-9448.2018.12.037
[32] 陈静, 张尚暖. 铁死亡相关基因DPP4与肿瘤免疫及预后的生物信息学研究[J]. 中国免疫学杂志,2023,39(11):2348−2354,2360. [CHEN Jing, ZHANG Shangnuan. Bioinformatics study on ferroptosis related gene DPP4 in tumor immunity and prognosis[J]. Chinese Journal of Immunology,2023,39(11):2348−2354,2360.] doi: 10.3969/j.issn.1000-484X.2023.11.018 CHEN Jing, ZHANG Shangnuan. Bioinformatics study on ferroptosis related gene DPP4 in tumor immunity and prognosis[J]. Chinese Journal of Immunology, 2023, 39(11): 2348−2354,2360. doi: 10.3969/j.issn.1000-484X.2023.11.018
[33] 胡太平, 曹建国. 芹菜素诱导人胃癌细胞凋亡作用及机制研究[J]. 国际病理科学与临床杂志,2007,27(1):6−10,93. [HU Taiping, CAO Jianguo. Study on pro-apoptotic effect of apigenin on human gastric cancer cells and its underlying mechanisms[J]. International Journal of Pathology and Clinical Medicine,2007,27(1):6−10,93.] HU Taiping, CAO Jianguo. Study on pro-apoptotic effect of apigenin on human gastric cancer cells and its underlying mechanisms[J]. International Journal of Pathology and Clinical Medicine, 2007, 27(1): 6−10,93.
[34] 肖静. 乙酰胆碱酯酶在癌症基因—病毒靶向治疗系统中的抗癌作用[D]. 新乡:河南师范大学, 2014. [XIAO Jing. The anti-tumor effect of acetyl cholinesterase in cacer gene-viro targeting therapy system[D]. Xinxiang:Henan Normal University, 2014.] XIAO Jing. The anti-tumor effect of acetyl cholinesterase in cacer gene-viro targeting therapy system[D]. Xinxiang: Henan Normal University, 2014.
[35] 张中惠. 神经干细胞调控过程中相关神经分子的定量分析[D]. 上海:华东师范大学, 2022. [ZHANG Zhonghui. Quantitative analysis of ralated neuralmolecules in the regulation of neural stem cells[D]. Shanghai:East China Normal University, 2022.] ZHANG Zhonghui. Quantitative analysis of ralated neuralmolecules in the regulation of neural stem cells[D]. Shanghai: East China Normal University, 2022.
[36] 安楠. 运动对成长期大鼠神经肌肉接头发育的影响及机制研究[D]. 北京:北京体育大学, 2009. [AN Nan. Effects and mechanism of activities on neuromuscular junction development in young rats[D]. Beijing:Beijing Sport University, 2009.] AN Nan. Effects and mechanism of activities on neuromuscular junction development in young rats[D]. Beijing: Beijing Sport University, 2009.
[37] 徐恩斌, 张忠兵, 谢渭芬. 乙酰胆碱酯酶的研究进展[J]. 国外医学(生理、病理科学与临床分册),2003,23(1):73−75. [XU Enbin, ZHANG Zhongbing, XIE Weifen. Research progress of acetylcholinesterase[J]. Foreign Medical Sciences (Section of Pathophysiology and Clinical Medicine),2003,23(1):73−75.] XU Enbin, ZHANG Zhongbing, XIE Weifen. Research progress of acetylcholinesterase[J]. Foreign Medical Sciences (Section of Pathophysiology and Clinical Medicine), 2003, 23(1): 73−75.





 下载:
下载:
 下载:
下载:



