Biological Characteristics Analysis of Limosilactobacillus reuteri JBR3 and the Effect of Protective Agents on Its Viability
-
摘要: 为推进罗伊氏粘液乳杆菌IMAUJBR3在肉类发酵剂中的应用,对该菌株进行了全基因组、代谢系统、生物学特性分析,同时采用单因素和响应面试验分析获得乳酸菌冷冻干燥的最佳保护剂比例。结果表明,JBR3的基因组由1个环状染色体和2个环状质粒组成,在代谢系统中注释到gene0325、gene1042等5个产甾醇酯酶(EC 3.1.1.13)基因,该酶可以水解生成甾醇和脂肪酸。在益生特性分析中,未注释到生物胺相关基因,但发现了groEL、groES等多个伴侣蛋白基因和冷应激基因gene0733和gene1677,这些基因的存在确保乳酸菌在冷应激期间仍然可以保证菌株的功能特性不被破坏,说明该菌株适合于冷冻干燥。同时,JBR3包含完整的NADH系统、yfeX过氧化物酶、gor谷胱甘肽还原酶、trxA硫氧还蛋白和trxB硫氧还蛋白还原酶,JBR3可能具有较好的抗氧化能力,可以作为发酵剂应用于发酵食品中。JBR3在脱脂乳粉为9.3 g/100 mL、麦芽糊精为2.3 g/100 mL、谷氨酸钠6.8 g/100 mL、海藻糖为10.0 g/100 mL的保护剂复配下冷冻干燥,存活率最高为95.17%±0.57%,活菌数为9.20×1010 CFU/g,该菌株具有良好的应用前景。Abstract: In order to advance the application of Limosilactobacillus reuteri IMAUJBR3 in meat fermenters, the strain was subjected to comprehensive genomic, metabolic and biological analyses. Additionally, one-way and response surface experiments were conducted to identify the optimal ratio of protectants for freeze-drying of lactic acid bacteria. The genome of JBR3 was found to consist of one circular chromosome and two circular plasmids. Five sterol esterase-producing enzyme (EC 3.1.1.13) genes, including gene0325 and gene1042, were identified in the metabolic system, which could be hydrolysed to produce sterols and fatty acids. In the probiotic characterisation, no biogenic amine-related genes were annotated. However, several chaperonin genes, such as groEL and groES, and cold stress genes, gene0733 and gene1677 were found. The presence of these genes ensured that the functional properties of the strain could be maintained during the period of cold stress in lactic acid bacteria. This suggested that the strain was suitable for freeze-drying. In addition, JBR3 contained an intact NADH system, yfeX peroxidase, gor glutathione reductase, trxA thioredoxin and trxB thioredoxin reductase. This suggested that JBR3 might have a superior antioxidant capacity, which could be exploited as a fermentation agent in fermented foods. JBR3 was freeze-dried under a protective agent complex of skimmed milk powder (9.3 g/100 mL), maltodextrin (2.3 g/100 mL), monosodium glutamate (6.8 g/100 mL), and trehalose (10.0 g/100 mL). The survival rate of the strain was 95.17%±0.57%, with a viable bacterial number of 9.20×1010 CFU/g. The strain has a promising future in various applications.
-
Keywords:
- Limosilactobacillus reuteri /
- whole genome /
- freeze-drying /
- protective agent /
- survival rate
-
罗伊氏粘液乳杆菌(Limosilactobacillus reuteri)属于厚壁菌门(Firmicutes),芽孢杆菌纲(Bacilli),乳杆菌目(Lactobacillales),乳杆菌科(Lactobacillaceae),乳酸杆菌属(Lactobacillus),属于革兰氏阳性兼性厌氧菌。2003年被国家卫生部批准可作为人类保健品的微生物菌种,也是国际上公认的新型益生乳酸菌,已广泛应用于食品发酵行业。其也是已报道几乎天然存在于所有脊椎动物和哺乳动物肠道内的乳酸菌,是美国FDA(Food and Drug Administration)认可的一种安全健康的食品补充剂[1]。它经常被添加在各种发酵食品、食品补充剂或婴儿配方奶粉中[2]。研究表明,罗伊粘液乳杆菌具有肠粘膜屏障功能,可通过促进粘液蛋白分泌,增加肠粘膜厚度,促进肠粘膜发育,从而改善肠道形态结构[3];还可以通过产生罗伊氏菌素,具有明显的抑菌能力和改善肠道菌群或预防治疗肠道感染的功能[4−5]。但目前的研究主要集中于罗伊氏粘液乳杆菌对人类益生效果的影响,对于其作为发酵剂对肉制品的影响还未受到足够的关注。
从市场销售和工业应用情况来看,现在乳酸菌大部分都是以干燥之后的固态粉末状形式存在。粉末状与液体相比运输方便、质量更容易控制、总体的成本低[6]。与喷雾干燥相比较,冷冻干燥制得乳酸菌发酵剂活菌数量较高、遗传性能稳定、活力好,储藏和运输方便[7−8]。尽管冷冻干燥已成为保存细菌细胞最常用的方法[9],但是它仍然会对乳酸菌细胞造成一定的伤害,这些伤害主要表现在冷冻干燥过程中细胞内外部形成冰晶,冰晶对细胞产生机械损伤影响细胞膜的完整性[10−11]。冷冻干燥过程中,磷脂的极性端除去水合氢键,迫使酰基集聚在一起,极性端密度和糖链间的范德华力增加,液晶相脂膜转变凝胶相,增加细胞膜的透过性,在细胞复水后胞内的细胞质、离子、蛋白质和酶等物质溶出,从而导致细胞功能丢失、活力下降甚至死亡等[12−13]。Chen等[14]研究NaHCO3、MgSO4、抗坏血酸钠、谷氨酸钠、磷酸盐缓冲液分别对冷冻干燥过程中保加利亚乳杆菌的生存影响,发现谷氨酸钠和磷酸盐缓冲液对存活率的影响更大,说明添加不同类物质对乳酸菌的保护效果不同。保护剂的成分复杂,不同的菌株对冻干的反应特性不尽相同,所以通过单因素和响应面试验筛选出最优的保护剂复配比例,可提高罗伊氏粘液乳杆菌的活性。
本研究以从中国内蒙古传统风干羊肉制品分离得到具有低温发酵能力和高产有机酸能力的罗伊氏粘液乳杆菌IMAUJBR3为研究对象,对JBR3进行全基因、代谢系统、生物性特性分析,以便更好地了解罗伊氏粘液乳杆菌JBR3的功能特性。同时进行单因素和响应面试验优化冷冻干燥过程中的最优保护剂比例,对推动我国肉制品专用发酵剂工业化生产具有重要的意义。
1. 材料与方法
1.1 材料与仪器
罗伊氏粘液乳杆菌IMAUJBR3(JBR3) 内蒙古农业大学肉品科学与技术团队从中国内蒙古传统风干羊肉制品分离获得,于−80 ℃进行保存,保藏号:CGMCC NO.29998,NCBI登陆号:SAMN38259820;脱脂乳粉 内蒙古伊利实业集团股份有限公司;麦芽糖糊精、海藻糖、谷氨酸钠 河南万邦化工科技有限公司;MRS培养基 北京索莱宝科技有限公司;细菌总DNA提取试剂盒 天根生化科技(北京)有限公司。
Alpha 1-4 LSC Baisc真空冷冻干燥机 德国Christ公司;Synergy酶标仪 德国Biotek;Centrifuge 5810R高速离心机 德国Eppendorf;TM4000扫描电镜 日本日立;BCV-2FD超净台、LHS-100CA恒温培养箱 上海一恒科学仪器有限公司。
1.2 实验方法
1.2.1 菌株培养及菌体的获取
将待测菌株JBR3接入到MRS液体培养基中,20 ℃培养18~24 h,使活菌数达到107 CFU/mL。将活化好的乳酸菌接入到MRS液体培养基中20 ℃培养18~24 h,使活菌数达到108 CFU/mL,连续传代2次获得待测菌液。
将待测菌液接入至50 mL离心管中,4000 r/min离心10 min,倒掉上清,加入0.85%无菌生理盐水振匀,4000 r/min离心10 min,重复上述操作3次,获得菌泥[15]。
1.2.2 全基因组测序
使用细菌总DNA提取试剂盒对罗伊氏粘液乳杆菌JBR3的DNA进行提取。将提取的DNA以27F、1492R为引物进行PCR扩增,将扩增样品送至上海美吉生物医药科技有限公司,采用Illumina+PacBio的测序方式进行测序。将全基因测序结果上传至上海美吉生物的I-Sanger云平台(www.i-sanger.com),利用云平台对数据进行分析。
1.2.3 单因素实验
以无菌水作为对照组(0 g/100 mL),以1 g菌泥加入1 mL保护剂的比例,按照表1分别向菌泥中加入相应浓度的保护剂。将含有保护剂的样品置于−80 ℃的冰箱预冻12 h。然后将其置于冷冻干燥机中进行冷冻干燥,程序为:冷冻干燥机预冻30 min,温度−80 ℃,预冻结束后进行冷冻干燥,条件为:温度−85 ℃,压力0.08 mbar,冻干时间26 h。考察不同浓度保护剂对乳酸菌存活率的影响。
表 1 单因素保护剂浓度Table 1. Single-factor protective agent concentration保护剂 浓度(g/100 mL) 脱脂乳粉 6、8、10、12、14 麦芽糊精 1、3、5、7、9 海藻糖 2、4、6、8、10 谷氨酸钠 1、3、5、7、9 1.2.4 响应面试验
结合单因素实验结果,依据Box-Behnken试验设计原则[16],得到脱脂乳粉、麦芽糊精、谷氨酸钠和海藻糖四个因素对菌株存活率影响较大的保护剂添加量,各取3个水平分别记为水平−1、0、1,进行因素水平共29个实验点的响应面分析试验,并运用Design Expert 11软件进行数据分析[17],进而得到最佳菌株存活率的条件,4因素3水平响应面试验设计表2。
表 2 响应面试验设计Table 2. Surface analysis experimental design因素 水平 −1 0 1 A脱脂乳粉(g/100 mL) 8 10 12 B麦芽糊精(g/100 mL) 1 3 5 C谷氨酸钠(g/100 mL) 5 7 9 D海藻糖(g/100 mL) 6 8 10 1.2.5 冷冻干燥存活率计算
将冷冻干燥后的菌粉用MRS培养基复水至冷冻干燥前相同体积,用平板计数法测定冷冻干燥前后活菌数。重复3次实验,每次3个平行。存活率计算公式[18]如下:
存活率(%)=NANB×100 式中:NA为冷冻干燥后的活菌数;NB为冷冻干燥前的活菌数。
1.2.6 扫描电镜观察形态
使用扫描电镜对冷冻干燥后的菌粉进行观察。将导电胶带贴于样品台中心,将干燥后的菌粉均匀粘在导电胶带上,吹去浮粉。将样品室抽真空后使用3.0 kV加速电压进行扫描成像。
1.3 数据处理
使用Box-Behnken进行响应面试验设计,利用Design Expert 11进行响应面试验数据统计分析。利用Office Excel 2019进行数据整理,利用Origin 2021进行绘图及IMB SPSS 26进行显著性分析。
2. 结果与分析
2.1 罗伊氏粘液乳杆菌JBR3基因组特性分析及功能注释
2.1.1 JBR3基因组特征分析
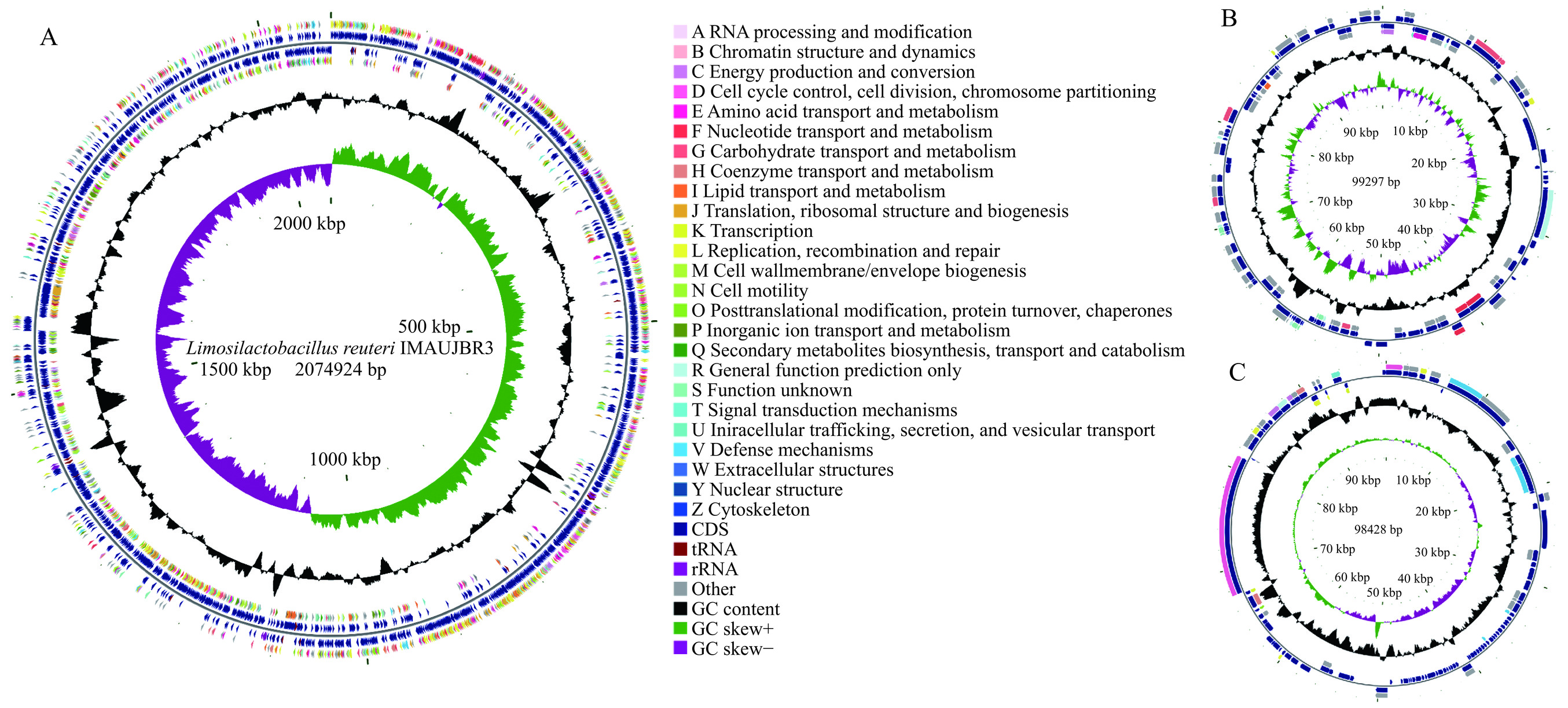
罗伊氏粘液乳杆菌JBR3含有一条环状染色体和两个环状质粒[19],结果如图1所示。全基因组大小为2074924 bp,平均G+C含量为39.17%,编码区共有2236个基因,平均长度为875 bp(表3)。质粒A基因组大小为99297 bp,平均G+C含量为38.33%,质粒B基因组大小为98428,平均G+C含量为38.29%,两条质粒均为New-Plasmid。在编码基因中,有2028个编码基因位于染色体上,平均长度887 bp,占整个基因组长度的86.72%;109个编码基因位于染色体A上,平均长度727 bp,99个编码基因位于染色体B上,平均长度780 bp。该基因组包含70个tRNAs,18个rRNAs,18个串联重复序列,其中11个串联重复序列在质粒B上,7个在染色体上,质粒A无串联重复序列。
![]() 图 1 罗伊氏粘液乳杆菌JBR3基因圈图注:图A表示为罗伊氏粘液乳杆菌IMAUJBR3的一条染色体的基因圈图;图B表示为罗伊氏粘液乳杆菌IMAUJBR3质粒A的基因圈图;图C表示为罗伊氏粘液乳杆菌IMAUJBR3质粒B的基因圈图。圈图从外到内第一圈和第四圈为正链、负链上的CDS,不同的颜色表示不同的COG功能分类;第二圈和第三圈分别为正链、负链上的CDS、tRNA、rRNA;第五圈为GC含量,向外的部分表示该区域GC含量高于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大,向内的部分表示该区域GC含量低于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大;第六圈为GC-Skew值,具体算法为(G−C)/(G+C),可以辅助判断前导链和后滞链,一般前导链GC skew>0,后滞链GC skew<0,也可以辅助判断复制起点(累计偏移最小值)和终点(累计偏移最大值),尤其对环状基因组最为重要;最内一圈为基因组大小标识。Figure 1. Gene circle map of Limosilactobacillus reuteri JBR3表 3 罗伊氏粘液乳杆菌IMAUJBR3基因组特征Table 3. Characteristics of the Limosilactobacillus reuteri IMAUJBR3 genome
图 1 罗伊氏粘液乳杆菌JBR3基因圈图注:图A表示为罗伊氏粘液乳杆菌IMAUJBR3的一条染色体的基因圈图;图B表示为罗伊氏粘液乳杆菌IMAUJBR3质粒A的基因圈图;图C表示为罗伊氏粘液乳杆菌IMAUJBR3质粒B的基因圈图。圈图从外到内第一圈和第四圈为正链、负链上的CDS,不同的颜色表示不同的COG功能分类;第二圈和第三圈分别为正链、负链上的CDS、tRNA、rRNA;第五圈为GC含量,向外的部分表示该区域GC含量高于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大,向内的部分表示该区域GC含量低于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大;第六圈为GC-Skew值,具体算法为(G−C)/(G+C),可以辅助判断前导链和后滞链,一般前导链GC skew>0,后滞链GC skew<0,也可以辅助判断复制起点(累计偏移最小值)和终点(累计偏移最大值),尤其对环状基因组最为重要;最内一圈为基因组大小标识。Figure 1. Gene circle map of Limosilactobacillus reuteri JBR3表 3 罗伊氏粘液乳杆菌IMAUJBR3基因组特征Table 3. Characteristics of the Limosilactobacillus reuteri IMAUJBR3 genome属性 值 属性 值 基因组大小(bp) 2074924 rRNAs 18 G+C含量(%) 39.17 5S rRNA 6 编码基因数 2236 16S rRNA 6 编码基因平均长度(bp) 875 23S rRNA 6 COGs基因数量 1844 tRNAs 70 质粒 2 sRNA 125 重复序列 18 2.1.2 罗伊氏粘液乳杆菌JBR3功能注释
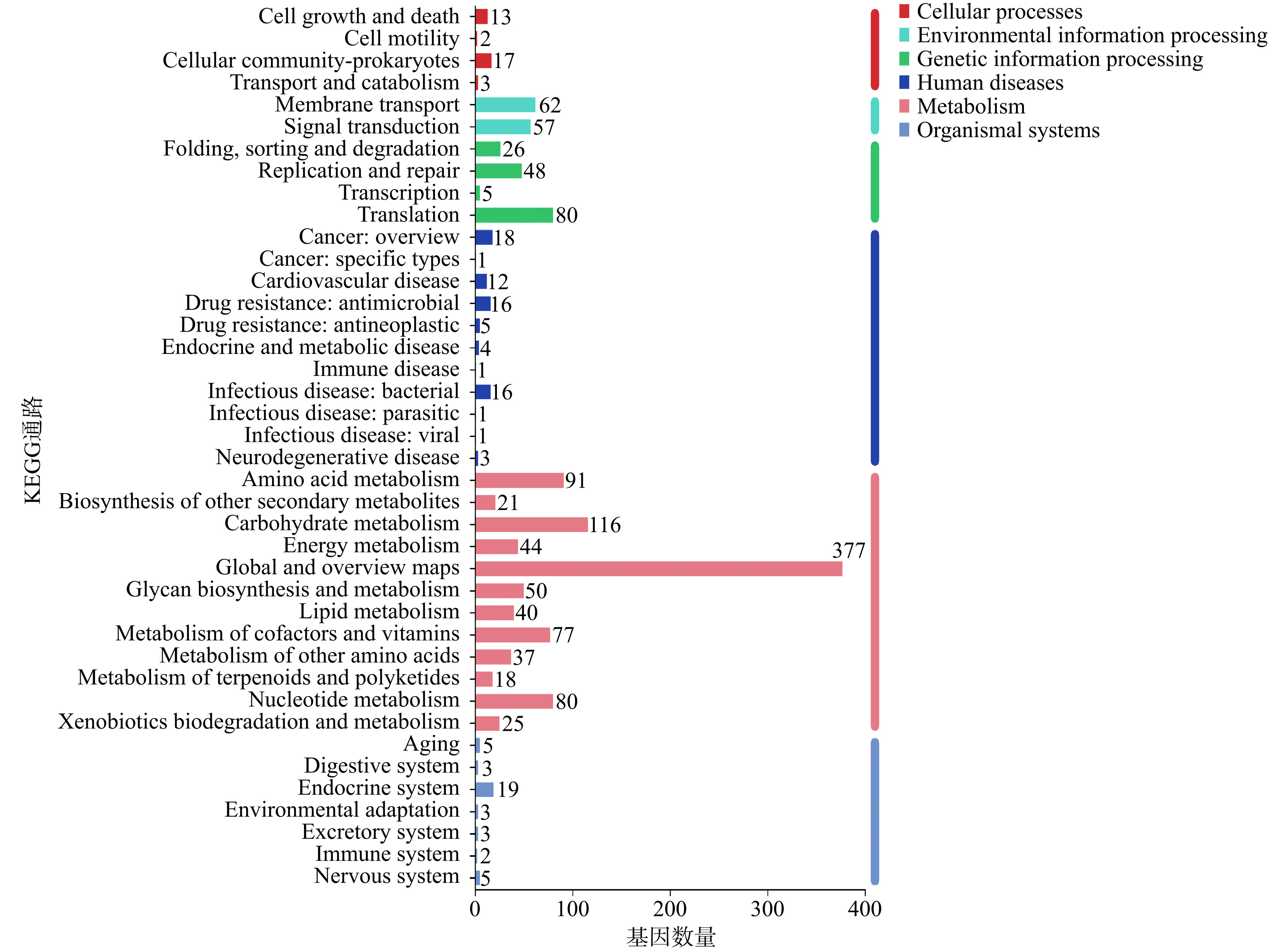
COG(Clusters of Orthologous Groups of proteins)功能注释显示,罗伊氏粘液乳杆菌JBR3的2236个蛋白编码基因中,有1844个(82.47%)基因被注释,共分为23个亚类和4个大类,包括Metabolism(新陈代谢)(34.78%)、Cellular processes and signaling(细胞过程和信号传递)(43.48%)、Information storage and processing(信息存储和处理)(13.04%)和特征不明显(Poorly characterized)(表4)。为了进一步阐明罗伊氏粘液乳杆菌JBR3基因功能,分析发现共注释了124个碳水化合物代谢基因,在碳水化合物活性酶中共注释到54种酶,碳水化合物酶11个、糖苷水解酶18个、糖基转移酶22个及辅助氧化还原酶3个。JBR3共有1379个(61.67%)基因被KEGG(Kyoto Encyclopedia of Genes and Genomes)注释,其中与新陈代谢(Metabolism)相关的基因有976个(70.78%),其次是遗传信息处理(Genetic Information Processing)188个(13.63%)(图2)。在JBR3的KEGG代谢注释中,氨基酸代谢(Amino acid metabolism)和碳水化合物代谢(Carbohydrate metabolism)被认为是该菌株的主要功能,分别含有91和116个基因。
表 4 罗伊氏粘液乳杆菌JBR3功能注释Table 4. Limosilactobacillus reuteri JBR3 functional notesCOG分类 COG功能描述 基因数量 细胞过程和信号传导
(Cellular processes and signaling)细胞周期控制、细胞分裂、染色体分配(Cell cycle control, cell division, chromosome partitioning) 38 细胞壁/膜/包被的生物发生(Cell wall/membrane/envelope biogenesis) 113 细胞运动(Cell motility) 16 翻译后修饰、蛋白质周转、合子(Posttranslational modification, protein turnover, chaperones) 65 信号转导机制(Signal transduction mechanisms) 78 细胞内贩运、分泌和囊泡运输(Intracellular trafficking, secretion, and vesicular transport) 15 防御机制(Defense mechanisms) 50 细胞外结构(Extracellular structures) 2 细胞骨架(Cytoskeleton) 3 移动体:噬菌体、转座子(Mobilome: prophages, transposons) 303 信息存储和处理
(Information storage and processing)翻译、核糖体结构和生物发生(Translation,ribosomal structure and biogenesis) 187 转录(Transcription) 115 复制、重组和修复(Replication, recombination and repair) 106 新陈代谢(Metabolism) 能量产生和转换(Energy production and conversion) 71 氨基酸转运和代谢(Amino acid transport and metabolism) 141 核苷酸转运和代谢(Nucleotide transport and metabolism) 103 碳水化合物的运输和代谢(Carbohydrate transport and metabolism) 124 辅酶运输和代谢(Coenzyme transport and metabolism) 94 脂质运输和代谢(Lipid transport and metabolism) 72 无机离子转运和代谢(Inorganic ion transport and metabolism) 70 次生代谢物的生物合成、运输和分解(Secondary metabolites biosynthesis, transport and catabolism) 17 特征不明显(Poorly characterized) 仅预测一般功能(General function prediction only) 151 功能未知(Function unknown) 78 2.2 罗伊氏粘液乳杆菌JBR3代谢系统分析
2.2.1 JBR3碳水化合物活性酶
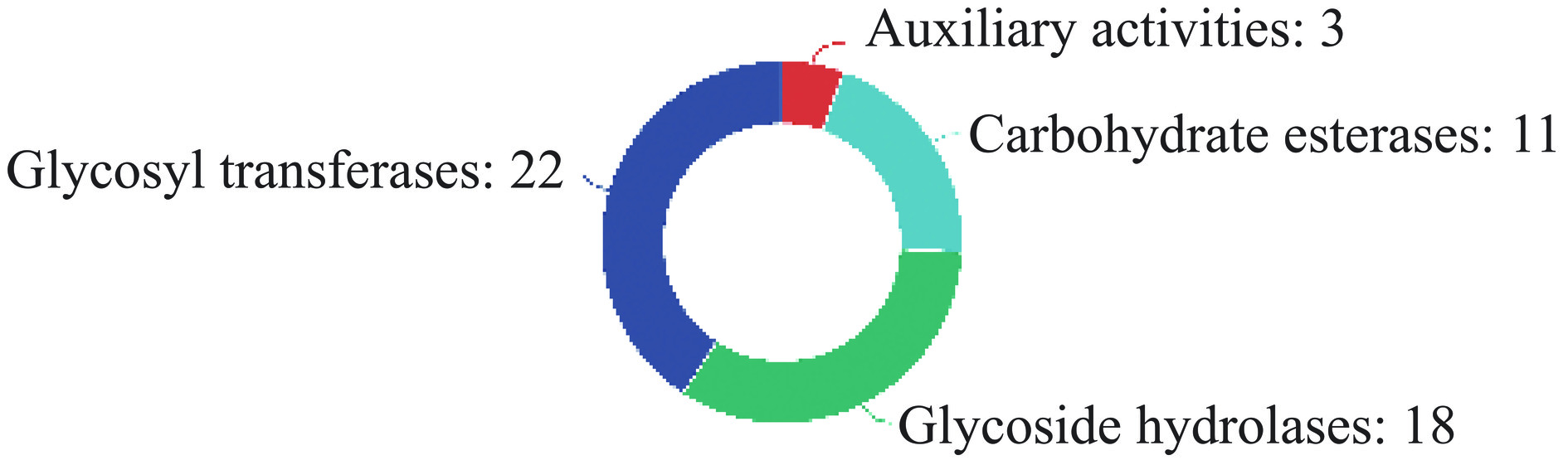
碳水化合物在很多生物学功能中具有重要地位,通过研究碳水化合物相关酶可以得到大量有意义的生物学信息。因此利用碳水化合物活性酶(Carbohydrate-active enzymes,CAZyme)数据库对罗伊氏粘液乳杆菌JBR3的碳水化合物活性酶进行注释(图3)。共注释54个基因,其中在染色体上的有51个,质粒A上有3个。辅助氧化还原酶(Auxiliary Activities,AAs)3个,碳水化合物酯酶(Carbohydrate Esterases,CEs)11个,糖苷水解酶(Glycoside Hydrolases,GHs)18个,糖基转移酶(Glycosyl Transferases,GTs)22个。在碳水化合物酯酶的基因中发现有5个基因(gene0325、gene1042、gene1698、gene1737和pA_gene0092)产生甾醇酯酶(EC 3.1.1.13),其中4个基因位于染色体上,1个基因位于质粒A上,甾醇酯酶(EC 3.1.1.13)属于CE10家族。
2.2.2 JBR3次级代谢产物合成分析
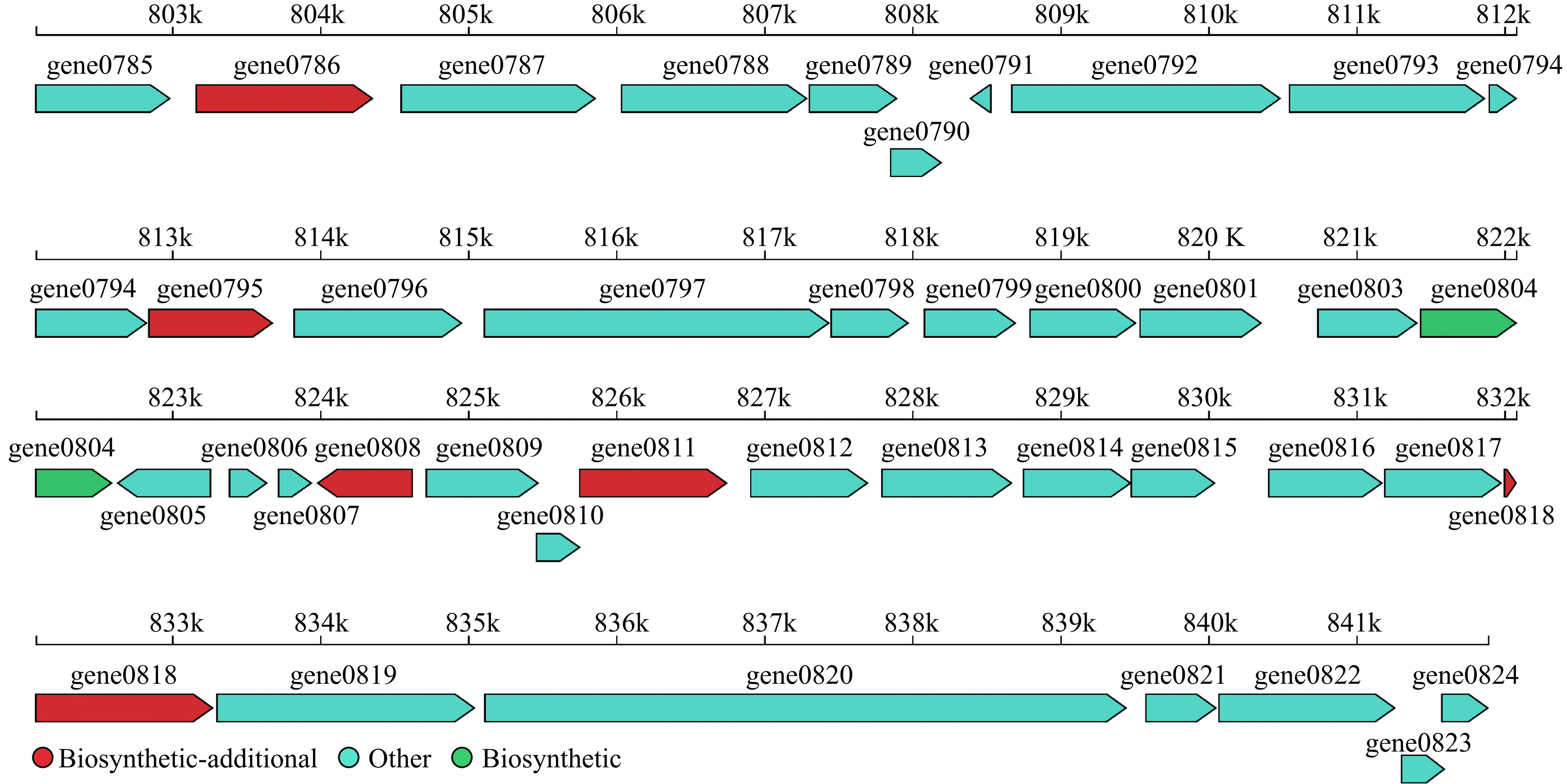
通过使用antiSMASH软件对样本的次级代谢产物合成基因簇进行预测(图4)。只鉴定出1个基因簇,基因簇的合成类型为T3PKS,长度为41158 bp,共有40个基因在这个基因簇上。只有6个基因被COG注释到了相应的功能,基因gene0804具有生物合成功能,gene0786、gene0795、gene0808、gene0811和gene0818被注释为附加生物合成基因,gene0795被注释为具有脱氢酶/还原酶家族氧化还原酶(SDR family oxidoreductase),这说明JBR3产生的代谢产物可能具有一定的抗氧化能力。其他34个基因未被注释到功能。
2.3 罗伊氏粘液乳杆菌JBR3功能特性评价
2.3.1 产生物胺基因评价
生物胺是一类广泛存在于发酵食品中的低分子量含氮有机化合物。它们可以由含有氨基酸脱羧酶的乳酸菌产生,该酶可以将氨基酸前体转化为生物胺。其中酪氨是发酵产品中含量最高的一种,常用作食品腐败/新鲜度的质量指标[20−21]。因此利用Blast技术在VFDB数据库中检索与酪胺(基因:tryS、tyrDC和tdcP)、组胺(基因:nhaC、hdcP、hdcA、hdcB和hisS)、腐胺(基因:speF和potE)和尸胺(基因:cadA、cadB和cadC)相关的基因。在VFDB数据库中并没有检索到与产生生物胺相关的氨基酸脱羧酶完整基因簇。JBR3未检索到产生物胺的常见的基因,作为肉制品发酵剂更为安全。
2.3.2 环境适应性及应激相关基因分析
罗伊氏粘液乳杆菌是人类和动物胃肠道中乳杆菌属的主要物种之一,具有更好的环境适应性及应激能力和较强的耐热、耐胁迫等能力[22]。在基因组中发现了许多适应性应激反应基因,如伴侣蛋白groEL、CLP酶等。基因groES、groEL、hslO、clpC和clpE等已经被证明与热应激有一定的关系(表5)[23]。同时罗伊氏粘液乳杆菌JBR3也检测到cspA基因,这个基因与冷应激有一定关系。同时还发现了dnaK和dnaJ基因,dnaK和dnaJ存在可以防止应激期间蛋白质在细胞内聚集,保证乳酸菌在应激期间的功能特性不被破坏[24]。同时还含有一个aspS基因,该基因可能会产生碱性编码胁迫蛋白,该蛋白的产生可以让乳酸菌更好地定植于肠道,减少胃肠道中的酸和胆汁盐对益生菌存活及活力的威胁[25]。JBR3基因中还包含atpA、atpH等编码完整的质子泵F0F1 ATPase系统和可能的Na+/H+反转运蛋白,它们是酸胁迫反应的关键蛋白。它们可以通过消耗细胞内的三磷酸腺苷将H+泵出细胞外,以维持细胞内的pH稳定,并适应酸性环境或酸胁迫[25]。通过检索发现JBR3具有较好的环境适应性及应激相关基因,并筛选出了与热应激有关的基因9个,耐冷基因2个,还有一些适应性伴侣蛋白等,这表明JBR3可以通过冷冻干燥的方式制备发酵剂。
表 5 益生特性评价相关基因Table 5. Evaluation of genes related to probiotic properties序号 基因名称 基因功能描述 基因ID 1 – 通用应激蛋白家族 gene0572、gene1309、gene1310、
gene1683、gene1923、gene18812 groEL 伴侣蛋白 gene0401 3 clpL ATP依赖性Clp蛋白酶、ATP结合亚基 gene0044 4 clpP ATP依赖性Clp内肽酶蛋白、水解亚基ClpP gene0443 5 clpX ATP依赖性Clp蛋白酶、ATP结合亚基 gene0788 6 clpP Clp蛋白酶 gene1372 7 clpE ATP依赖性Clp蛋白酶、ATP结合亚基 gene1438 8 clpP Clp蛋白酶 gene1590 9 clpC ATP依赖性Clp蛋白酶、ATP结合亚基 gene1661 10 groES 共同伴侣GroES gene0400 11 groEL 共同伴侣GroEL gene0401 12 hslO Hsp33家族分子伴侣HslO gene0286 13 clpC ATP依赖性Clp蛋白酶、ATP结合亚基 gene1661 14 clpE ATP依赖性Clp蛋白酶、ATP结合亚基 gene1438 15 hrcA 热诱导转录抑制因子 gene0830 16 – 热休克蛋白 gene1329 17 dnaK 伴侣蛋白DnaK gene0832 18 dnaJ 伴侣蛋白DnaK gene0833 19 aspS 天冬氨酸-tRNA连接酶 gene0853 20 cspA 冷休克蛋白 gene0733、gene1677 21 atpH ATP合酶F1亚基δ gene0544 22 atpB F0F1 ATP合酶亚基A gene0541 23 atpE F0F1 ATP合酶亚基C gene0542 24 atpF F0F1 ATP合酶亚基B gene0543 25 atpA F0F1 ATP合酶亚基α gene0545 26 atpG F0F1 ATP合酶亚基γ gene0546 27 atpD F0F1 ATP合酶亚基β gene0547 28 atpC F0F1 ATP合成酶亚基ε gene0548 29 nhaC Na+/H+反转运体家族 gene0169、gene1984 30 – Na+/H+交换器家族 gene0213、gene1122 31 – NADH氧化酶 gene1818 32 yfeX Dyp型过氧化物酶 gene1735 33 trxA 硫氧还蛋白 gene0627、gene1978 34 trxB 硫氧还蛋白 gene0427 35 trxB 硫氧还蛋白还原酶 gene1125 36 gor 谷胱甘肽还原酶 gene1532、pA_gene0001 37 gor – pA_gene0109 注:表中基因名称一列中“–”表示还未被命名的基因名称,基因功能描述一列中“–”表示检测到的基因名称未被注释到相关信息。 2.3.3 抗氧化能力
在肉制品发酵过程,乳酸菌的存在不仅可以改变发酵肉制品的风味,提高营养价值,还可以减少机体在肠道中活性氧自由基(Reactive oxygen species,ROS)的积累,提高抗氧化活性,帮助人体预防由于氧化引发的多种疾病[26−27]。因此对罗伊氏粘液乳杆菌JBR3全基因组抗氧化能力进行评估(表5),发现罗伊氏粘液乳杆菌JBR3基因组具有完整的NADH(Nicotinamide adenine dinucleotide)系统(NADH氧化酶和NADH过氧化物酶)、过氧化物酶、谷胱甘肽还原酶、硫氧还蛋白和硫氧还蛋白还原酶。同时发现JBR3的抗氧化基因中,有两个基因在质粒A上,其中有一个未被注释到相关功能。说明JBR3可能具有较好的抗氧化能力,可以作为发酵剂应用于发酵食品中。
2.4 单一保护剂对罗伊氏粘液乳杆菌JBR3冷冻干燥存活率的影响
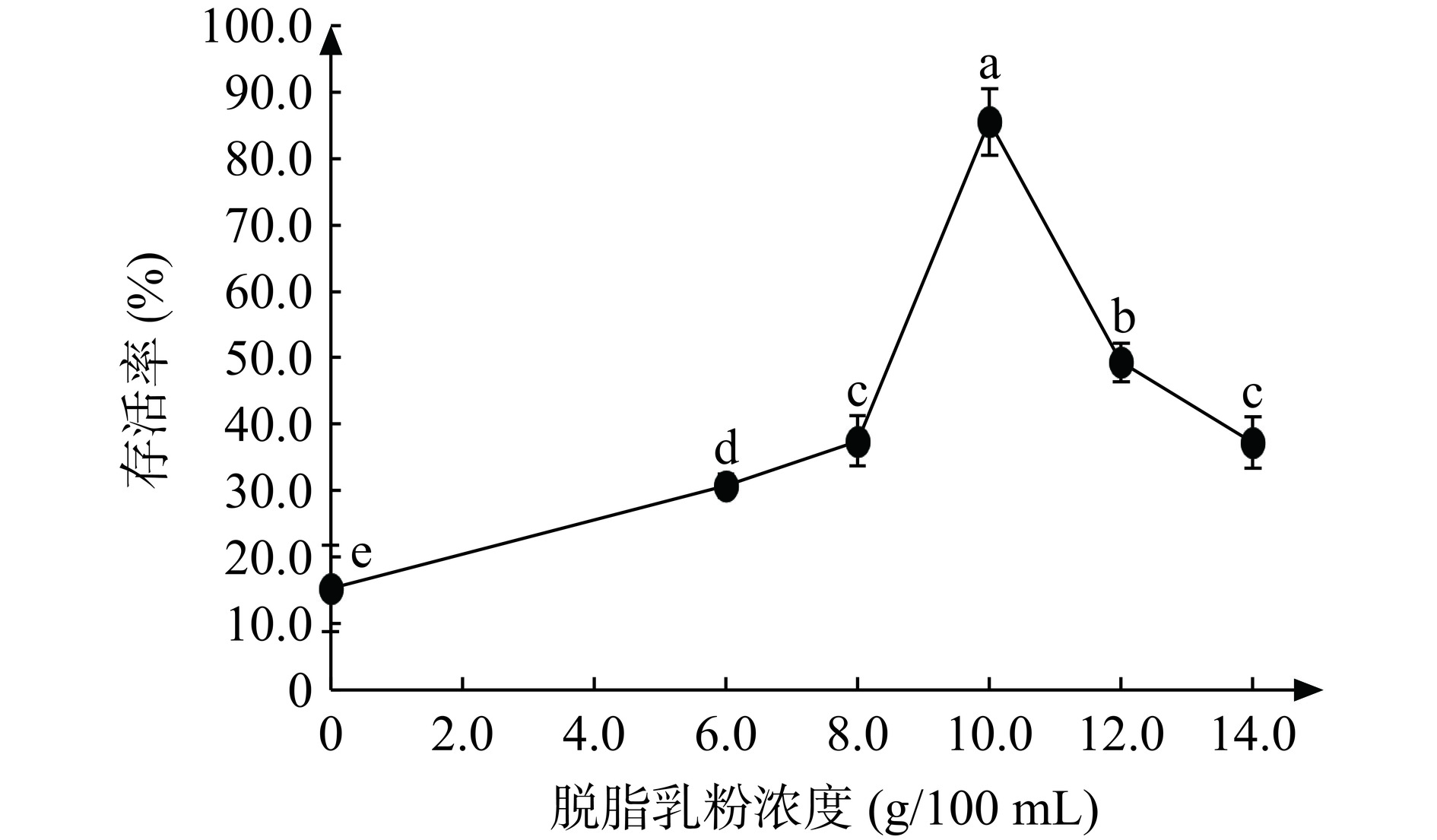
2.4.1 脱脂乳粉对JBR3冷冻干燥后存活率的影响
有研究表明脱脂乳粉可以将菌体“包裹”在内部,减少菌体与外部冰晶的接触,防止细胞膜发生泄露,减少物理损伤[28−29]。因此选用浓度为6、8、10、12、14 g/100 mL的脱脂乳粉作为保护剂,结果如图5。随着脱脂乳粉浓度增加,乳酸菌的存活率先增加后降低,这可能是因为随着脱脂乳粉浓度升高,粘性也相对增加,容易使细胞脱水,影响了细胞的形态,从而影响了菌体的存活率[30]。以0 g/100 mL的脱脂乳粉作为保护剂时,JBR3的存活率约为15.30%,活菌数约为1.48×1010 CFU/g。这可能是因为JBR3基因组中包含cspA、dnaK和dnaJ等多个胁迫蛋白基因,在冷冻干燥过程中防止蛋白质在细胞内聚集,保证乳酸菌在应激期间的功能特性不被破坏,使乳酸菌的活菌数可以维持在1010 CFU/g。当脱脂乳粉浓度在10 g/100 mL时,存活率最高为85.50%(活菌数8.27×1010 CFU/g),显著高于其他浓度下的存活率(P<0.05)。与对照组相比,脱脂乳粉冷冻干燥后的乳酸菌活菌数要高于对照组。说明脱脂乳粉的添加可以提高冷冻干燥后乳酸菌的存活率。因此选择脱脂乳粉浓度8、10、12 g/100 mL进行响应面试验。
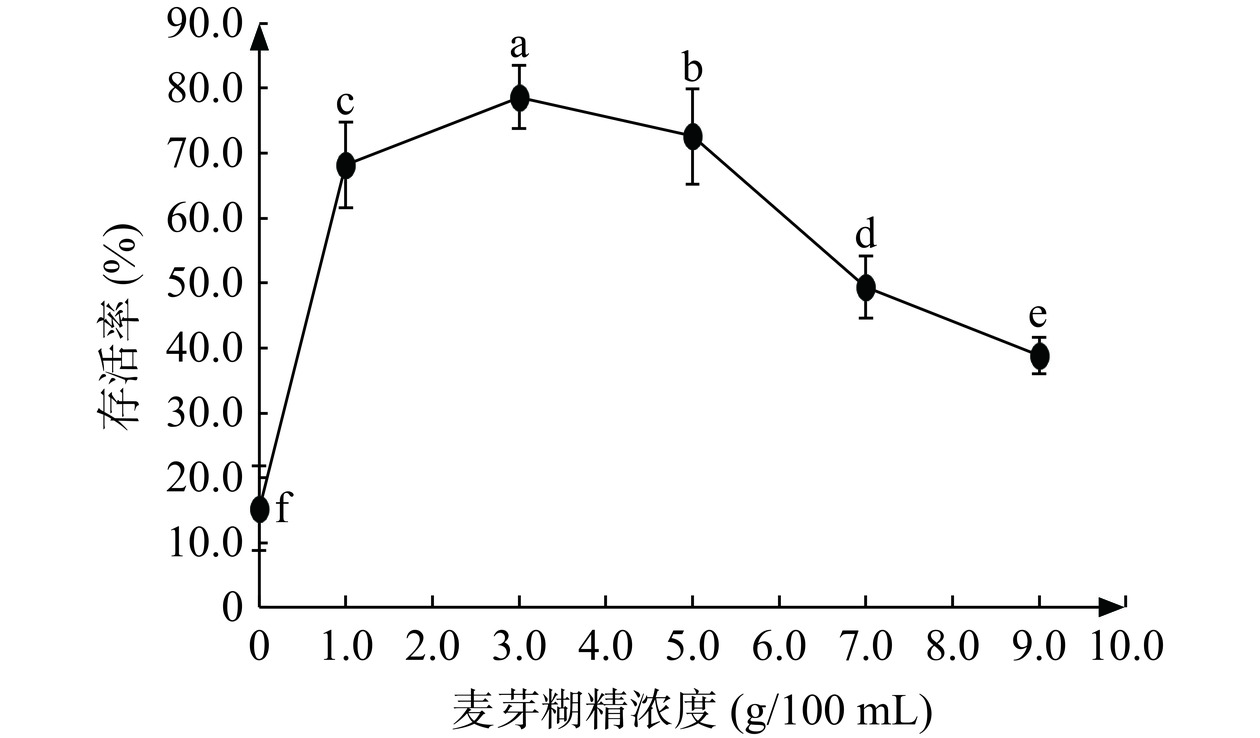
2.4.2 麦芽糊精对JBR3冷冻干燥后存活率的影响
接入不同浓度麦芽糊精对乳酸菌存活率的影响如图6所示。当麦芽糊精浓度为3 g/100 mL时,乳酸菌的存活率最高为78.62%(活菌数7.6×1010 CFU/g),当浓度继续增加后,乳酸菌存活率随之降低,这可能是高浓度麦芽糊精包裹了菌体细胞导致菌体丧失了自身保护的反应系统的启动[31],存活率均显著高于对照组(麦芽糊精浓度0 g/100 mL)(P<0.05),说明添加适量的麦芽糊精,可以提高冷冻干燥后乳酸菌的存活率。因此选择麦芽糊精浓度1、3、5 g/100 mL进行响应面试验。
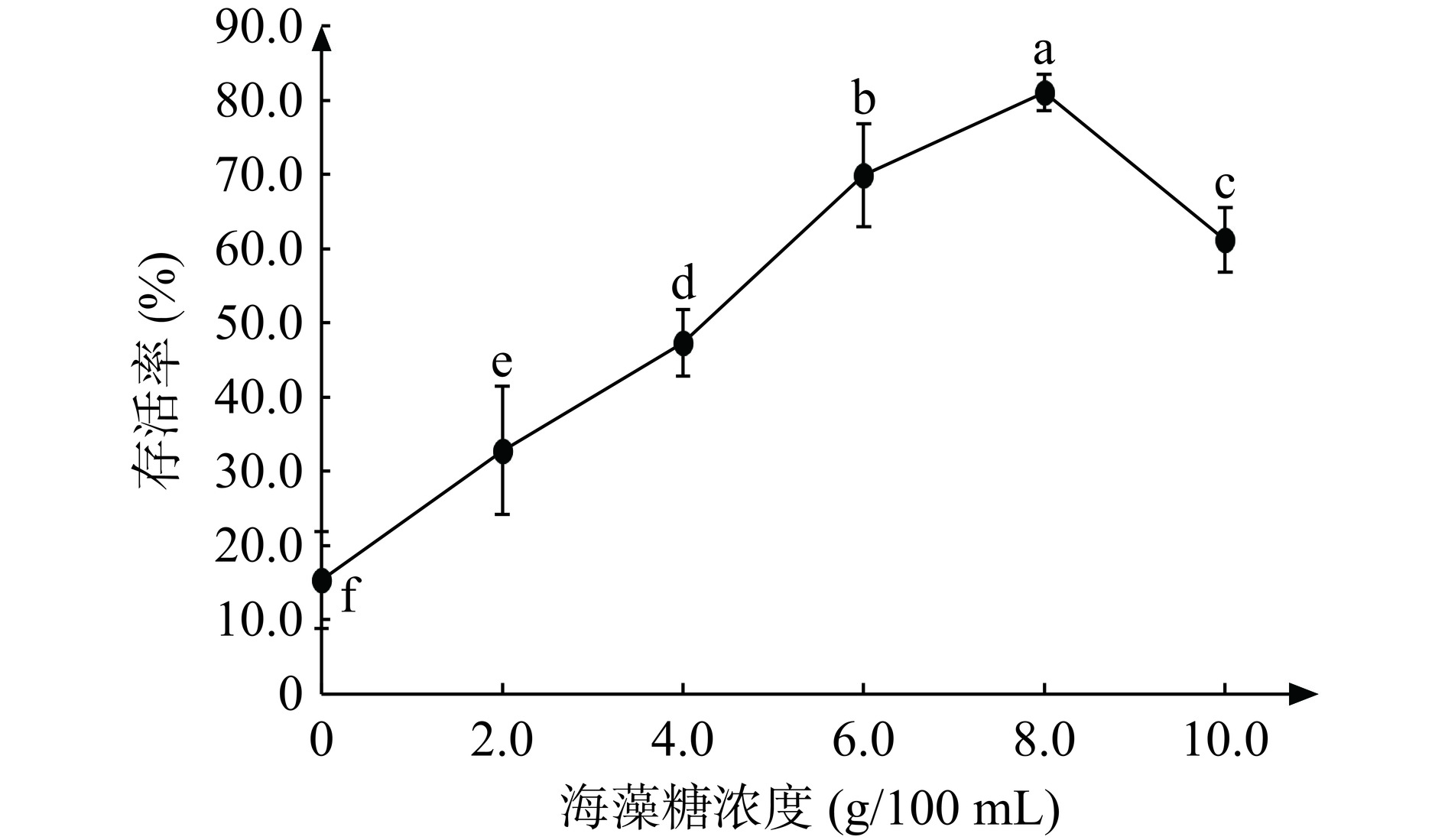
2.4.3 海藻糖对JBR3冷冻干燥后存活率的影响
糖类物质可以包裹着菌体,防止蛋白的延伸和胞内物质的泄露,保持菌体结构的稳定,起到保护菌体的作用[32−33]。添加不同浓度的海藻糖对冷冻干燥乳酸菌的存活率影响结果如图7。研究发现,海藻糖浓度为8 g/100 mL时,乳酸菌存活率最高为81.06%(活菌数7.84×1010 CFU/g)。在含有不同浓度海藻糖的乳酸菌中,随着浓度的增加,存活率先增加后降低,这可能是过高的海藻糖浓度引起细胞内外渗透压失衡,导致细胞脱水,降低了对细胞膜稳定性的维持效果[15]。这也与Cui等[34]研究表面物质对乳酸菌冷冻干燥存活的影响,发现在有海藻酸盐的保护下可以减少细菌表面物质的产生,乳酸菌存活率显著提高的现象较为一致。因此选择海藻糖浓度6、8、10 g/100 mL进行响应面试验。
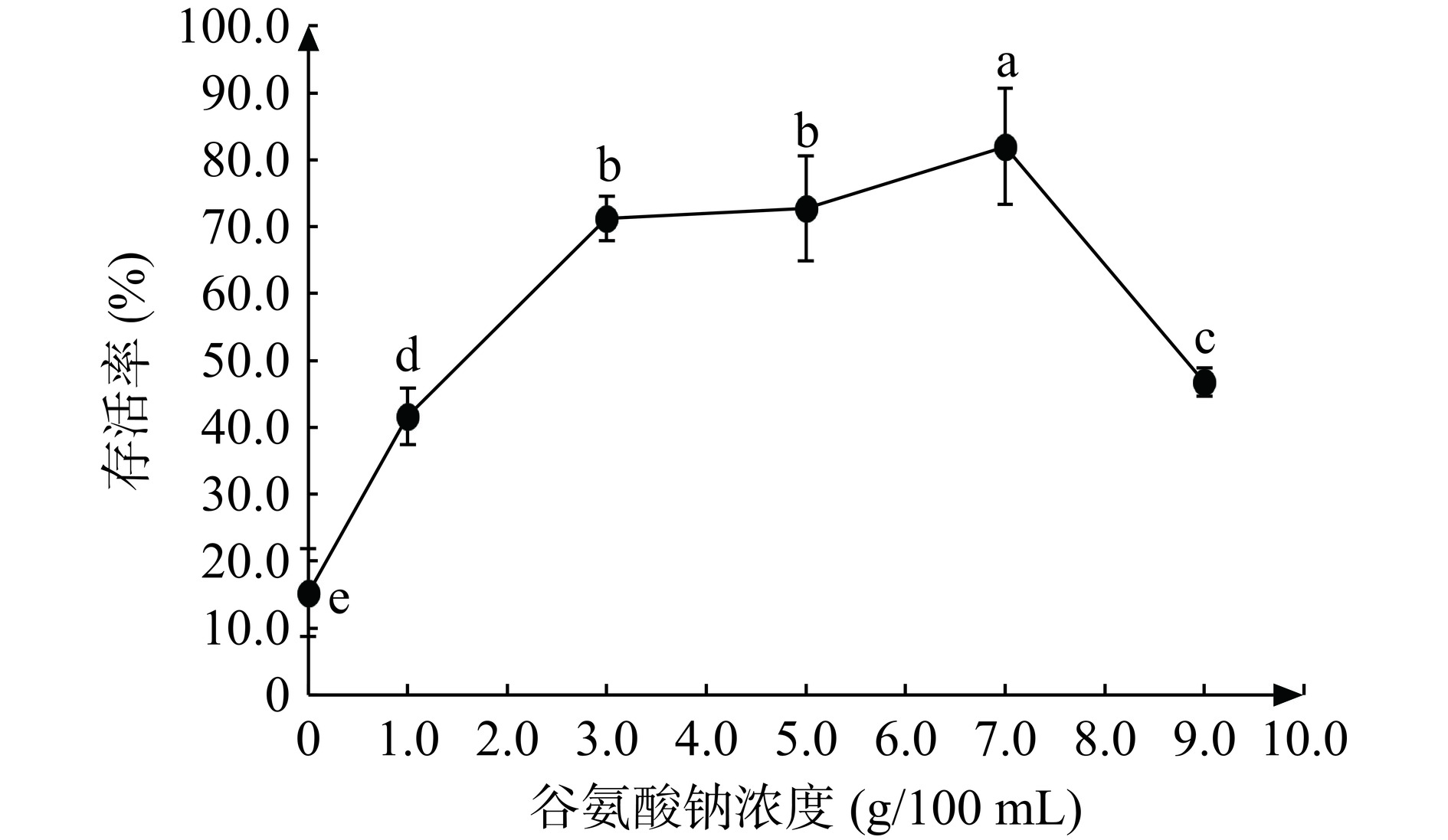
2.4.4 谷氨酸钠对JBR3冷冻干燥后存活率的影响
谷氨酸钠作为有机化合物和钠盐的一种,有机盐通过渗透作用进入细胞内部,提高细胞内部电解质溶液浓度,减少在冻干过程中的溶质损伤[35]。通过研究发现,当谷氨酸钠浓度为7 g/100 mL时,冷冻干燥后罗伊氏粘液乳杆菌JBR3存活率最高,为82.00%(活菌数7.90×1010 CFU/g)。加入过量的谷氨酸钠会导致菌体渗透压增加,降低了菌粉存活率[36]。因此选择谷氨酸钠浓度5、7、9 g/100 mL进行响应面试验。
2.5 复配保护剂对罗伊氏粘液乳杆菌JBR3冷冻干燥存活率的影响
2.5.1 响应面分析复配保护剂对JBR3冷冻干燥存活率的试验设计与结果
根据不同浓度保护剂对乳酸菌存活率影响的单因素实验结果,依据Box-Behnken试验设计原则,选择脱脂乳粉、麦芽糊精、谷氨酸钠和海藻糖对菌株存活率影响较大的3个水平,进行响应面分析。试验结果如表6。
表 6 响应面分析菌株存活率试验验设计与结果Table 6. Response surface analysis to optimise strain survival rate experimental design and results试验号 A脱脂乳粉
(g/100 mL)B麦芽糊精
(g/100 mL)C谷氨酸钠
(g/100 mL)D海藻糖
(g/100 mL)菌株存活
率(%)1 8 1 7 8 71.92 2 12 1 7 8 60.03 3 8 5 7 8 63.99 4 12 5 7 8 89.29 5 10 3 5 6 59.73 6 10 3 9 6 78.93 7 10 3 5 10 83.88 8 10 3 9 10 83.58 9 8 3 7 6 53.63 10 12 3 7 6 78.95 11 8 3 7 10 90.28 12 12 3 7 10 76.26 13 10 1 5 8 71.31 14 10 5 5 8 78.91 15 10 1 9 8 77.71 16 10 5 9 8 77.41 17 8 3 5 8 62.16 18 12 3 5 8 74.08 19 8 3 9 8 64.30 20 12 3 9 8 74.05 21 10 1 7 6 66.74 22 10 5 7 6 77.12 23 10 1 7 10 88.15 24 10 5 7 10 79.31 25 10 3 7 8 93.26 26 10 3 7 8 89.60 27 10 3 7 8 92.34 28 10 3 7 8 94.22 29 10 3 7 8 94.17 通过对实验数据进行二次回归方程分析、多元回归拟合,设脱脂乳粉、麦芽糊精、谷氨酸钠和海藻糖分别为A、B、C、D,以菌株存活率为响应值进行多元回归拟合,得到二次多项回归方程:乳酸菌存活率(%)=92.72+3.86A+2.51B+2.16C+7.2D+9.3AB−0.54AC−9.84AD−1.97BC−4.81BD−4.88CD−13.23A2−7.86B2−9.84C2−6.03D2。菌株存活率模型回归方差分析结果如表7所示。
表 7 菌株存活率模型及回归系数的回归分析结果Table 7. Results of regression analysis of strain survival rate model and regression coefficients来源 离差平方和 自由度 均方 F值 P值 显著性 模型 3510.55 14 250.75 22.22 <0.0001 ** A-脱脂乳粉 179.26 1 179.26 15.89 0.0014 ** B-麦芽糊精 75.85 1 75.85 6.72 0.0213 * C-谷氨酸钠 55.94 1 55.94 4.96 0.0429 * D-海藻糖 621.5 1 621.5 55.08 <0.0001 ** AB 345.77 1 345.77 30.64 <0.0001 ** AC 1.18 1 1.18 0.1 0.7515 AD 386.91 1 386.91 34.29 <0.0001 ** BC 15.6 1 15.6 1.38 0.2592 BD 92.35 1 92.35 8.18 0.0126 * CD 95.06 1 95.06 8.42 0.0116 * A2 1135.32 1 1135.32 100.62 <0.0001 ** B2 400.84 1 400.84 35.52 <0.0001 ** C2 628.2 1 628.2 55.67 <0.0001 ** D2 235.65 1 235.65 20.88 0.0004 ** 残差 157.97 14 11.28 失拟项 143.45 10 14.34 3.95 0.0988 不显著 纯误差 14.52 4 3.63 总和 3668.52 28 R2=0.9569 R2Adj=0.9139 注:**表示极显著,P<0.01,*表示显著,P<0.05。 从表7可以看出该回归模型极显著(P<0.01),失拟项不显著(P=0.0988>0.05),同时模型回归系数R2=0.9569,调节后的R2=0.9139,表明91.39%的数据可用该模型解释,说明方程可靠性较高,模型拟合程度好,实验值与预测值比较接近,因此可以用该模型对菌株存活率进行分析和预测。通过分析方差数据可以看出,一次项脱脂乳粉、海藻糖对菌株存活率具有极显著影响(P<0.01),谷氨酸钠、麦芽糊精对菌株存活率具有显著影响(P<0.05),分析各因素的主效应关系为:D>A>B>C,即海藻糖>脱脂乳粉>麦芽糊精>谷氨酸钠。其二次项响应面作用AB、AD对菌株存活率具有极显著的影响(P<0.01),BD、CD对菌株存活率具有显著的影响(P<0.05)。
2.5.2 复配保护剂的响应面优化分析
以菌株存活率最大值为优化目标,由Design-Expert 10.0.3软件对实验进行优化,结果如图9。得到预测菌株存活率为95.42%,4个因素的预测值分别是脱脂乳粉为9.28 g/100 mL、麦芽糊精为2.32 g/100 mL、谷氨酸钠为6.81 g/100 mL、海藻糖为10.0 g/100 mL,为了确定该模型的准确性,采用优化后的参数进行验证实验,为方便操作,条件参数设为脱脂乳粉为9.3 g/100 mL、麦芽糊精为2.3 g/100 mL、谷氨酸钠为6.8 g/100 mL、海藻糖为10.0 g/100 mL,该条件下重复实验3次,测得菌注存活率平均值为95.17%±0.57%,活菌数为9.20×1010 CFU/g,与模型预测值95.42%在5%偏差范围内,说明该模型优化得到的工艺参数可靠。同时,也有许多研究表明多种类型保护剂复配应用于乳酸菌冷冻干燥过程,可以显著提高乳酸菌冷冻干燥后的菌株活力。Cheng等[37]将10%脱脂牛奶、7%海藻糖、2%山梨醇和0.8%酪氨酸的混合液作为冷冻保护剂对植物乳植杆菌L1和L2进行冷冻干燥,冻干后乳酸菌的存活率较未添加保护剂的存活率显著提升,存活率均达到90%以上。Zhang等[38]在干酪乳杆菌Zhang冷冻干燥过程中添加谷胱甘肽作为保护剂,干酪乳杆菌Zhang的存活率显著提高到86.6%。Lee等[39]对植物乳杆菌JH287进行冷冻干燥,确定最佳保护培养基为10%山梨醇和10%脱脂乳,获得的细胞存活率为86.37%,略低于本实验的结果。这些研究结果表明乳酸菌冻干过程中复合保护剂可达到理想的保护效果,冻干后细胞的存活率可提高到85%以上。
2.6 保护剂对罗伊氏粘液乳杆菌JBR3菌粉微观结构的影响
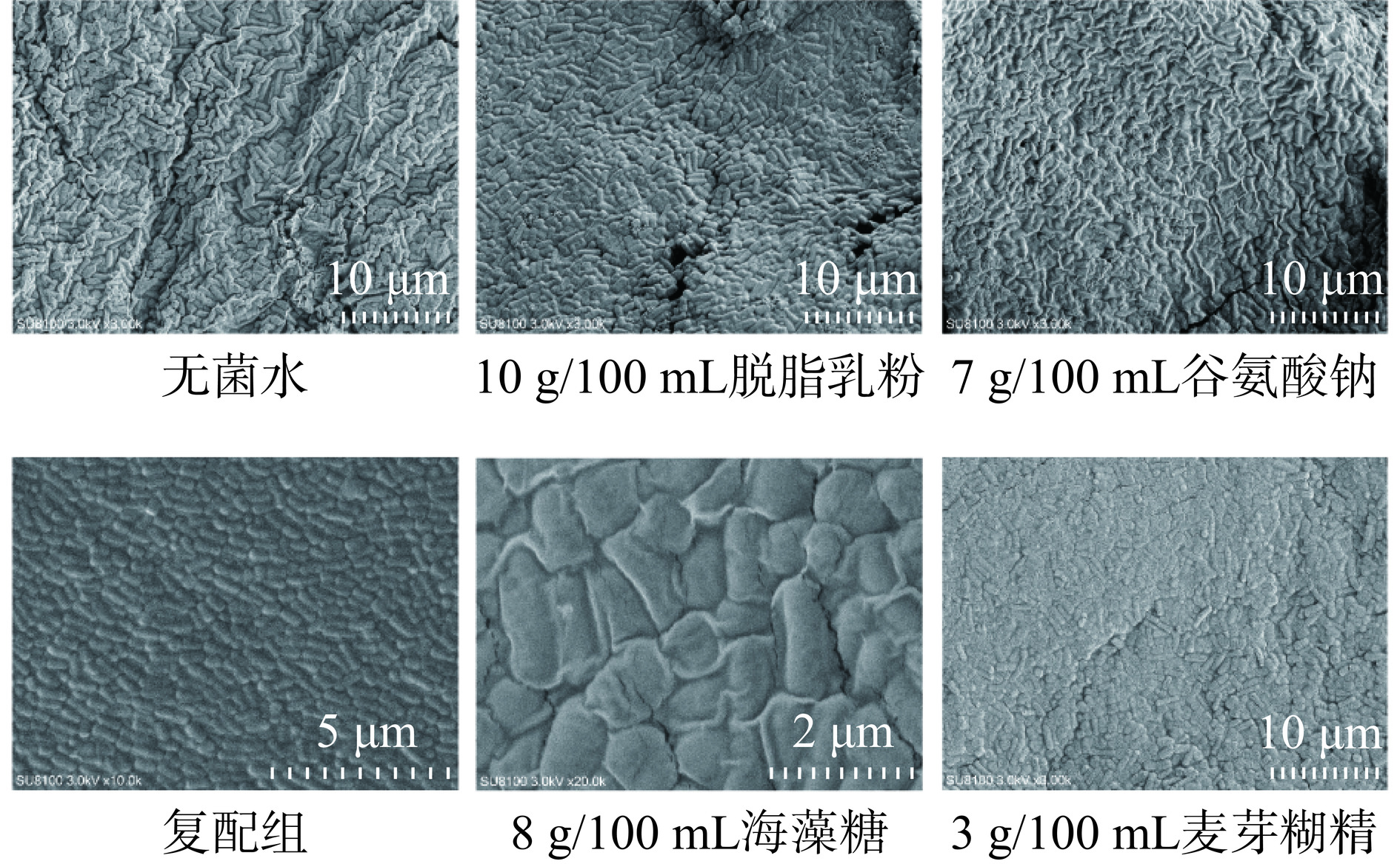
将最佳浓度保护剂加入JBR3菌泥中进行真空冷冻干燥,扫描电镜下观察菌粉微观结构,结果如图10所示。无菌水组、10 g/100 mL脱脂乳粉组、7 g/100 mL谷氨酸钠组和3 g/100 mL麦芽糊精组分别放大3000倍,8 g/100 mL海藻糖组放大20000倍。观察到添加了无菌水作为JBR3保护剂的菌粉菌体表面褶皱,且有菌体破裂导致胞内细胞质等物质溢出现象,这可能是在低温冷冻干燥过程中由于缺乏冻干保护剂的保护,菌体出现破裂损伤[40]。添加了8%海藻糖、10%脱脂乳粉、7%谷氨酸钠和3%麦芽糊精制备得到的JBR3冻干菌粉外表结构虽较无菌水组褶皱减少,但有较多孔洞,整体结构不稳定,甚至有部分细胞出现破裂、松散等现象。复配组放大10000倍,菌粉表面结构更加光滑平整,对菌体的包裹效果更全面,使得菌体结构均匀稳定,在冷冻干燥过程不易受到损伤,提高了乳酸菌JBR3冷冻干燥后的活性。可见复配保护剂明显提高了菌株对冷冻干燥的耐受能力。
3. 结论
JBR3基因组中包含1个环状染色体和2个环状质粒,共鉴定出2236个编码基因。注释到碳水化合物活性酶11个、糖苷水解酶18个、糖基转移酶22个及辅助氧化还原酶3个。JBR3包含完整的NADH系统,同时还注释到了dnaK、dnaJ等多个伴侣蛋白、适应性蛋白和2个冷胁迫蛋白(gene0733、gene1677),这些蛋白可以在冷冻干燥期间保护菌株的功能特性不被破坏。在KEGG中注释到包括糖酵解/糖异生代谢(ko00010)、氨基和核苷酸糖代谢(ko00520)和蔗糖和淀粉代谢(ko00500)等降解途径,这些代谢途径可以促进乳酸菌的新陈代谢,帮助细胞获得有机氧,进而促进氧化还原反应。在代谢系统分析中注释到gene0325、gene1042和gene1698等5个产生甾醇酯酶(EC 3.1.1.13)的基因,甾醇酯酶可以在水相条件下催化甾醇酯水解生成甾醇和脂肪酸,从而改变产品的风味物质。
为了更好地使用罗伊氏粘液乳杆菌JBR3,将其冷冻干燥制备成直投式发酵剂,通过对保护剂进行响应面试验,得到复配保护剂脱脂乳粉为9.3 g/100 mL、麦芽糊精为2.3 g/100 mL、谷氨酸钠6.8 g/100 mL、海藻糖为10.0 g/100 mL时,菌株存活率为95.17%±0.57%,活菌数为9.20×1010 CFU/g。JBR3的全基因组分析及发酵剂的制备可为罗伊氏粘液乳杆菌在后续的研究和应用提供理论基础。
-
图 1 罗伊氏粘液乳杆菌JBR3基因圈图
注:图A表示为罗伊氏粘液乳杆菌IMAUJBR3的一条染色体的基因圈图;图B表示为罗伊氏粘液乳杆菌IMAUJBR3质粒A的基因圈图;图C表示为罗伊氏粘液乳杆菌IMAUJBR3质粒B的基因圈图。圈图从外到内第一圈和第四圈为正链、负链上的CDS,不同的颜色表示不同的COG功能分类;第二圈和第三圈分别为正链、负链上的CDS、tRNA、rRNA;第五圈为GC含量,向外的部分表示该区域GC含量高于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大,向内的部分表示该区域GC含量低于全基因组平均GC含量,峰值越高表示与平均GC含量差值越大;第六圈为GC-Skew值,具体算法为(G−C)/(G+C),可以辅助判断前导链和后滞链,一般前导链GC skew>0,后滞链GC skew<0,也可以辅助判断复制起点(累计偏移最小值)和终点(累计偏移最大值),尤其对环状基因组最为重要;最内一圈为基因组大小标识。
Figure 1. Gene circle map of Limosilactobacillus reuteri JBR3
表 1 单因素保护剂浓度
Table 1 Single-factor protective agent concentration
保护剂 浓度(g/100 mL) 脱脂乳粉 6、8、10、12、14 麦芽糊精 1、3、5、7、9 海藻糖 2、4、6、8、10 谷氨酸钠 1、3、5、7、9 表 2 响应面试验设计
Table 2 Surface analysis experimental design
因素 水平 −1 0 1 A脱脂乳粉(g/100 mL) 8 10 12 B麦芽糊精(g/100 mL) 1 3 5 C谷氨酸钠(g/100 mL) 5 7 9 D海藻糖(g/100 mL) 6 8 10 表 3 罗伊氏粘液乳杆菌IMAUJBR3基因组特征
Table 3 Characteristics of the Limosilactobacillus reuteri IMAUJBR3 genome
属性 值 属性 值 基因组大小(bp) 2074924 rRNAs 18 G+C含量(%) 39.17 5S rRNA 6 编码基因数 2236 16S rRNA 6 编码基因平均长度(bp) 875 23S rRNA 6 COGs基因数量 1844 tRNAs 70 质粒 2 sRNA 125 重复序列 18 表 4 罗伊氏粘液乳杆菌JBR3功能注释
Table 4 Limosilactobacillus reuteri JBR3 functional notes
COG分类 COG功能描述 基因数量 细胞过程和信号传导
(Cellular processes and signaling)细胞周期控制、细胞分裂、染色体分配(Cell cycle control, cell division, chromosome partitioning) 38 细胞壁/膜/包被的生物发生(Cell wall/membrane/envelope biogenesis) 113 细胞运动(Cell motility) 16 翻译后修饰、蛋白质周转、合子(Posttranslational modification, protein turnover, chaperones) 65 信号转导机制(Signal transduction mechanisms) 78 细胞内贩运、分泌和囊泡运输(Intracellular trafficking, secretion, and vesicular transport) 15 防御机制(Defense mechanisms) 50 细胞外结构(Extracellular structures) 2 细胞骨架(Cytoskeleton) 3 移动体:噬菌体、转座子(Mobilome: prophages, transposons) 303 信息存储和处理
(Information storage and processing)翻译、核糖体结构和生物发生(Translation,ribosomal structure and biogenesis) 187 转录(Transcription) 115 复制、重组和修复(Replication, recombination and repair) 106 新陈代谢(Metabolism) 能量产生和转换(Energy production and conversion) 71 氨基酸转运和代谢(Amino acid transport and metabolism) 141 核苷酸转运和代谢(Nucleotide transport and metabolism) 103 碳水化合物的运输和代谢(Carbohydrate transport and metabolism) 124 辅酶运输和代谢(Coenzyme transport and metabolism) 94 脂质运输和代谢(Lipid transport and metabolism) 72 无机离子转运和代谢(Inorganic ion transport and metabolism) 70 次生代谢物的生物合成、运输和分解(Secondary metabolites biosynthesis, transport and catabolism) 17 特征不明显(Poorly characterized) 仅预测一般功能(General function prediction only) 151 功能未知(Function unknown) 78 表 5 益生特性评价相关基因
Table 5 Evaluation of genes related to probiotic properties
序号 基因名称 基因功能描述 基因ID 1 – 通用应激蛋白家族 gene0572、gene1309、gene1310、
gene1683、gene1923、gene18812 groEL 伴侣蛋白 gene0401 3 clpL ATP依赖性Clp蛋白酶、ATP结合亚基 gene0044 4 clpP ATP依赖性Clp内肽酶蛋白、水解亚基ClpP gene0443 5 clpX ATP依赖性Clp蛋白酶、ATP结合亚基 gene0788 6 clpP Clp蛋白酶 gene1372 7 clpE ATP依赖性Clp蛋白酶、ATP结合亚基 gene1438 8 clpP Clp蛋白酶 gene1590 9 clpC ATP依赖性Clp蛋白酶、ATP结合亚基 gene1661 10 groES 共同伴侣GroES gene0400 11 groEL 共同伴侣GroEL gene0401 12 hslO Hsp33家族分子伴侣HslO gene0286 13 clpC ATP依赖性Clp蛋白酶、ATP结合亚基 gene1661 14 clpE ATP依赖性Clp蛋白酶、ATP结合亚基 gene1438 15 hrcA 热诱导转录抑制因子 gene0830 16 – 热休克蛋白 gene1329 17 dnaK 伴侣蛋白DnaK gene0832 18 dnaJ 伴侣蛋白DnaK gene0833 19 aspS 天冬氨酸-tRNA连接酶 gene0853 20 cspA 冷休克蛋白 gene0733、gene1677 21 atpH ATP合酶F1亚基δ gene0544 22 atpB F0F1 ATP合酶亚基A gene0541 23 atpE F0F1 ATP合酶亚基C gene0542 24 atpF F0F1 ATP合酶亚基B gene0543 25 atpA F0F1 ATP合酶亚基α gene0545 26 atpG F0F1 ATP合酶亚基γ gene0546 27 atpD F0F1 ATP合酶亚基β gene0547 28 atpC F0F1 ATP合成酶亚基ε gene0548 29 nhaC Na+/H+反转运体家族 gene0169、gene1984 30 – Na+/H+交换器家族 gene0213、gene1122 31 – NADH氧化酶 gene1818 32 yfeX Dyp型过氧化物酶 gene1735 33 trxA 硫氧还蛋白 gene0627、gene1978 34 trxB 硫氧还蛋白 gene0427 35 trxB 硫氧还蛋白还原酶 gene1125 36 gor 谷胱甘肽还原酶 gene1532、pA_gene0001 37 gor – pA_gene0109 注:表中基因名称一列中“–”表示还未被命名的基因名称,基因功能描述一列中“–”表示检测到的基因名称未被注释到相关信息。 表 6 响应面分析菌株存活率试验验设计与结果
Table 6 Response surface analysis to optimise strain survival rate experimental design and results
试验号 A脱脂乳粉
(g/100 mL)B麦芽糊精
(g/100 mL)C谷氨酸钠
(g/100 mL)D海藻糖
(g/100 mL)菌株存活
率(%)1 8 1 7 8 71.92 2 12 1 7 8 60.03 3 8 5 7 8 63.99 4 12 5 7 8 89.29 5 10 3 5 6 59.73 6 10 3 9 6 78.93 7 10 3 5 10 83.88 8 10 3 9 10 83.58 9 8 3 7 6 53.63 10 12 3 7 6 78.95 11 8 3 7 10 90.28 12 12 3 7 10 76.26 13 10 1 5 8 71.31 14 10 5 5 8 78.91 15 10 1 9 8 77.71 16 10 5 9 8 77.41 17 8 3 5 8 62.16 18 12 3 5 8 74.08 19 8 3 9 8 64.30 20 12 3 9 8 74.05 21 10 1 7 6 66.74 22 10 5 7 6 77.12 23 10 1 7 10 88.15 24 10 5 7 10 79.31 25 10 3 7 8 93.26 26 10 3 7 8 89.60 27 10 3 7 8 92.34 28 10 3 7 8 94.22 29 10 3 7 8 94.17 表 7 菌株存活率模型及回归系数的回归分析结果
Table 7 Results of regression analysis of strain survival rate model and regression coefficients
来源 离差平方和 自由度 均方 F值 P值 显著性 模型 3510.55 14 250.75 22.22 <0.0001 ** A-脱脂乳粉 179.26 1 179.26 15.89 0.0014 ** B-麦芽糊精 75.85 1 75.85 6.72 0.0213 * C-谷氨酸钠 55.94 1 55.94 4.96 0.0429 * D-海藻糖 621.5 1 621.5 55.08 <0.0001 ** AB 345.77 1 345.77 30.64 <0.0001 ** AC 1.18 1 1.18 0.1 0.7515 AD 386.91 1 386.91 34.29 <0.0001 ** BC 15.6 1 15.6 1.38 0.2592 BD 92.35 1 92.35 8.18 0.0126 * CD 95.06 1 95.06 8.42 0.0116 * A2 1135.32 1 1135.32 100.62 <0.0001 ** B2 400.84 1 400.84 35.52 <0.0001 ** C2 628.2 1 628.2 55.67 <0.0001 ** D2 235.65 1 235.65 20.88 0.0004 ** 残差 157.97 14 11.28 失拟项 143.45 10 14.34 3.95 0.0988 不显著 纯误差 14.52 4 3.63 总和 3668.52 28 R2=0.9569 R2Adj=0.9139 注:**表示极显著,P<0.01,*表示显著,P<0.05。 -
[1] 李正华. 罗伊氏乳杆菌生物学特性及功能性发酵乳的研究[D]. 无锡:江南大学, 2008. [LI Z H. Studies on the biological properties of Lactobacillus reuteri and function fermented milk[D]. Wuxi:Jiangnan University, 2008.] LI Z H. Studies on the biological properties of Lactobacillus reuteri and function fermented milk[D]. Wuxi: Jiangnan University, 2008.
[2] DIEZ E P, MARTIN C I, GARRIDO M J, et al. Probiotic and functional properties of Limosilactobacillus reuteri INIA P572[J]. Nutrients, Basel:Mdpi,2021,13(6):1860. doi: 10.3390/nu13061860
[3] YI H B, WANG L, XIONG Y X, et al. Lactobacillus reuteri LR1 improved expression of genes of tight junction proteins via the MLCK pathway in IPEC-1 cells during infection with enterotoxigenic Escherichia coli K88[J]. Mediators of Inflammation,2018,2018:1−8.
[4] ROOS S, JONSSON H. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components[J]. Microbiology (Reading, England),2002,148:433−442. doi: 10.1099/00221287-148-2-433
[5] SPINLER J K, TAWEECHOTIPATR M, ROGNERUD C L, et al. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens[J]. Anaerobe,2008,14(3):166−171. doi: 10.1016/j.anaerobe.2008.02.001
[6] FU N, CHEN X D. Towards a maximl cell survival in convective thermal drying processes[J]. Food Research International,2011,44(5):1127−1149. doi: 10.1016/j.foodres.2011.03.053
[7] PEIREN J, BUYSE J, DE V P, et al. Improving survival and storage stability of bacteria recalcitrant to freeze-drying:A coordinated study by european culture collections[J]. Applied Microbiology and Biotechnology,2015,99(8):3559−3571. doi: 10.1007/s00253-015-6476-6
[8] VELLY H, BOUIX M, PASSOT S, et al. Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp. lactis TOMSC161[J]. Applied Microbiology and Biotechnology,2015,99(2):907−918. doi: 10.1007/s00253-014-6152-2
[9] MING L C, RAHIM R A, WAN H Y, et al. Formulation of protective agents for improvement of Lactobacillus salivarius I 24 survival rate subjected to freeze drying for production of live cells in powderized form[J]. Food & Bioprocess Technology,2009,2(4):431−436.
[10] MORGAN C A, HERMAN N, WHITE P A, et al. Preservation of micro-organisms by drying:A review[J]. Journal of Microbiological Methods,2006,66(2):183−193. doi: 10.1016/j.mimet.2006.02.017
[11] HELBER E B L, MAURICIO E R, LORENZO Z, et al. Effect of ethylene and 1-methylcyclopropene on the postharvest behavior of cape gooseberry fruits (Physalis peruviana L.)[J]. Food Science and Technology International,2017,23(1):86−96. doi: 10.1177/1082013216658581
[12] LESLIE S B, ISRAELI E, LIGHTHART B, et al. Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying[J]. Applied and Environmental Microbiology,1995,61(10):3592−3597. doi: 10.1128/aem.61.10.3592-3597.1995
[13] HOEKSTR A F, WILLEM F W, JULIA B, et al. Membrane stabilization in the dry state[J]. Comparative Biochemistry and Physiology-Part A:Physiology,1996,117(3):335−341.
[14] CHEN H, WANG J, LUO Q, et al. Effect of NaHCO3, MgSO4, sodium ascorbate, sodium glutamate, phosphate buffer on survival of Lactobacillus bulgaricus during freeze-drying[J]. Advance Journal of Food Science & Technology,2013,5(6):771−774.
[15] 张曦予, 李锐定, 莫明规, 等. 基于人工神经网络耦联遗传算法(BP-GA)优化干酪乳杆菌LTL1361冻干保护剂配方[J]. 食品工业科技,2022,43(21):175−184. [ZHANG X Y, LI R D, MO M G, et al. Optimization of lyophilized protective agent formulation of Lactobacillus casei LTL1361 based on artificial neural network coupled genetic algorithm (BP-GA)[J]. Science and Technology of Food Industry,2022,43(21):175−184.] ZHANG X Y, LI R D, MO M G, et al. Optimization of lyophilized protective agent formulation of Lactobacillus casei LTL1361 based on artificial neural network coupled genetic algorithm (BP-GA)[J]. Science and Technology of Food Industry, 2022, 43(21): 175−184.
[16] 陈红斌, 洪伦波, 汪娟, 等. 响应面优化高速剪切法制备薄荷挥发油包合物工艺[J]. 食品工业科技,2023,44(22):210−218. [CHEN H B, HONG L B, WANG J, et al. Preparation of peppermint oil/β-CD inclusion by high-speed shearing with process optimization using response surface methodology[J]. Science and Technology of Food Industry,2023,44(22):210−218.] CHEN H B, HONG L B, WANG J, et al. Preparation of peppermint oil/β-CD inclusion by high-speed shearing with process optimization using response surface methodology[J]. Science and Technology of Food Industry, 2023, 44(22): 210−218.
[17] 徐鹏飞, 王昊乾, 郑新飞, 等. 喷雾冷冻干燥技术制备乳双歧杆菌Probio-M8微胶囊制剂[J]. 中国食品学报,2021,21(7):197−207. [XU P F, WANG H Q, ZHENG X F, et al. Preparation of Bifidobacterium lactis Probio-M8 microcapsule by spray-freeze-drying technology[J]. Journal of Chinese Institute of Food Science and Technology,2021,21(7):197−207.] XU P F, WANG H Q, ZHENG X F, et al. Preparation of Bifidobacterium lactis Probio-M8 microcapsule by spray-freeze-drying technology[J]. Journal of Chinese Institute of Food Science and Technology, 2021, 21(7): 197−207.
[18] 周中凯, 王俊轩, 李莹. 青香蕉淀粉对乳酸菌在冻干和贮存过程中活性的保护效果[J]. 食品科学,2016,37(15):144−148. [ZHOU Z K, WANG J X, LI Y. Protective effect of unripe banana starch on viability of Lactobacillus plantarum during freeze-drying and storage[J]. Food Science,2016,37(15):144−148.] doi: 10.7506/spkx1002-6630-201615024 ZHOU Z K, WANG J X, LI Y. Protective effect of unripe banana starch on viability of Lactobacillus plantarum during freeze-drying and storage[J]. Food Science, 2016, 37(15): 144−148. doi: 10.7506/spkx1002-6630-201615024
[19] ZHANG L, ZHONG J, LIU H, et al. Complete genome sequence of the drought resistance-promoting endophyte Klebsiella sp. LTGPAF-6F[J]. Journal of Biotechnology,2017,246:36−39. doi: 10.1016/j.jbiotec.2017.02.008
[20] LI Y, HSIEH C H, LAI C W, et al. Tyramine detection using PEDOT:PSS/AuNPs/1-methyl-4-mercaptopyridine modified screen-printed carbon electrode with molecularly imprinted polymer solid phase extraction[J]. Biosensors & Bioelectronics,2017,87:142−149.
[21] WOJNOWSKI W, NAMIEśNIK J, JUSTYNA P W. Dispersive liquid-liquid microextraction combined with gas chromatography-mass spectrometry for in situ determination of biogenic amines in meat:Estimation of meat's freshness[J]. Microchemical Journal,2019,145:130−138. doi: 10.1016/j.microc.2018.10.034
[22] MA X Y, WOO J S, CHO J H, et al. IL-6 expression-suppressing Lactobacillus reuteri strains alleviate gut microbiota-induced anxiety and depression in mice[J]. Letters in Applied Microbiology,2023,77(1):1.
[23] KONSTANTINOS P, ÁNGEL A, BRON P A, et al. Stress physiology of lactic acid bacteria[J]. Microbiology and Molecular Biology Reviews:MMBR,2016,80(3):837−90. doi: 10.1128/MMBR.00076-15
[24] ANDREA M A, ERIC A, JUN Y G, et al. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism[J]. Applied and Environmental Microbiology,2008,74(15):4610−4625. doi: 10.1128/AEM.00054-08
[25] SANDERSON I R. The physicochemical environment of the neonatal intestine[J]. American Journal of Clinical Nutrition,1999,69(5):1028s−1034s. doi: 10.1093/ajcn/69.5.1028s
[26] WANG M, LEI M, SAMINA N, et al. Impact of Lactobacillus plantarum 423 fermentation on the antioxidant activity and flavor properties of rice bran and wheat bran[J]. Food Chemistry,2020,330:127156. doi: 10.1016/j.foodchem.2020.127156
[27] LIN X, XIA Y, YANG Y, et al. Probiotic characteristics of Lactobacillus plantarum AR113 and its molecular mechanism of antioxidant[J]. LWT,2020,126:109278. doi: 10.1016/j.lwt.2020.109278
[28] KHEM S, BANSAL V, SMALL D M, et al. Comparative influence of pH and heat on whey protein isolate in protecting Lactobacillus plantarum A17 during spray drying[J]. Food Hydrocolloids,2016,54:162−169. doi: 10.1016/j.foodhyd.2015.09.029
[29] KIM M, NAM D G, KIM S B, et al. Enhancement of viability, acid, and bile tolerance and accelerated stability in lyophilized Weissella cibaria JW15 with protective agents[J]. Food Science & Nutrition,2018,6(7):1904−1913.
[30] 李大鹏, 高玉荣. 益生菌植物乳杆菌G1-28复合冻干发酵剂制备及保藏条件研究[J]. 食品工业科技,2021,42(10):100−104. [LI D P, GAO Y R. Preparation and preservation technology of probiotic Lactobacillus plantarum G1-28 composite lyophilized starter[J]. Science and Technology of Food Industry,2021,42(10):100−104.] LI D P, GAO Y R. Preparation and preservation technology of probiotic Lactobacillus plantarum G1-28 composite lyophilized starter[J]. Science and Technology of Food Industry, 2021, 42(10): 100−104.
[31] 张涛, 闫有利. 草鱼肠道拮抗性芽孢杆菌的冻干保护剂优化研究[J]. 水产科学,2018,37(2):244−248. [ZHANG T, YAN Y L. Optimization of freeze-dried protective agent for pathogenic antagonistic bacillus in grass carp gut[J]. Fisheries Science,2018,37(2):244−248.] ZHANG T, YAN Y L. Optimization of freeze-dried protective agent for pathogenic antagonistic bacillus in grass carp gut[J]. Fisheries Science, 2018, 37(2): 244−248.
[32] SANTIVARANGKNA C, HIGL B, FOERST P. Protection mechanisms of sugars during different stages of preparation process of dried lactic acid starter cultures[J]. Food Microbiology,2007,25(3):429−441.
[33] MARINA G, VERA C, MAURICIO S, et al. Use of whey permeate containing in situ synthesised galacto-oligosaccharides for the growth and preservation of Lactobacillus plantarum[J]. Journal of Dairy Research,2013,80(3):374−381. doi: 10.1017/S0022029913000356
[34] CUI S M, PAN Z Y, WU S, et al. Improvement of the lyophilization survival rate of Lactobacillus casei via regulation of its surface substances[J]. Foods,2022,11(21):3468. doi: 10.3390/foods11213468
[35] WANG R X, SUN R Y, YANG Y, et al. Effects of salt stress on the freeze-drying survival rate of Lactiplantibacillus plantarum LIP-1[J]. Food Microbiology,2022,105:104009. doi: 10.1016/j.fm.2022.104009
[36] 张传伟, 朱慧霞, 柴佳太, 等. 低聚木糖复配冻干保护剂的优化及其对双歧杆菌微胶囊性能的影响[J]. 食品工业科技,2022,43(21):245−251. [ZHANG C W, ZHU H X, CHAI J T, et al. Optimization of cryoprotectants complexed with xylooligosaccharides and its effects on the properties of bifidobacterium microcapsules[J]. Science and Technology of Food Industry,2022,43(21):245−251.] ZHANG C W, ZHU H X, CHAI J T, et al. Optimization of cryoprotectants complexed with xylooligosaccharides and its effects on the properties of bifidobacterium microcapsules[J]. Science and Technology of Food Industry, 2022, 43(21): 245−251.
[37] CHENG Z Y, YAN X, WU J Y, et al. Effects of freeze drying in complex lyoprotectants on the survival, and membrane fatty acid composition of Lactobacillus plantarum L1 and Lactobacillus fermentum L2[J]. Cryobiology,2022,4(105):1−9.
[38] ZHANG J, LIU Q, CHEN W, et al. Short communication:Protection of lyophilized milk starter Lactobacillus casei Zhang by glutathione[J]. Journal of Dairy Science,2016,99(3):1846−1852. doi: 10.3168/jds.2015-9540
[39] LEE S B, KIM D H, PARK H D. Effects of protectant and rehydration conditions on the survival rate and malolactic fermentation efficiency of freeze-dried Lactobacillus plantarum JH287[J]. Applied Microbiology and Biotechnology,2016,100(18):7853−7863. doi: 10.1007/s00253-016-7509-5
[40] 刘开文, 马雯, 金刚. 真空冷冻干燥提高乳酸菌存活率及延长贮藏期的研究进展[J]. 食品科学,2024,45(2):325−333. [LIU K W, MA W, JIN G. Research progress on improving the survival rate and extending the storage period of lactic acid bacteria through vacuum freeze-drying[J]. Food Science,2024,45(2):325−333.] doi: 10.7506/spkx1002-6630-20230801-002 LIU K W, MA W, JIN G. Research progress on improving the survival rate and extending the storage period of lactic acid bacteria through vacuum freeze-drying[J]. Food Science, 2024, 45(2): 325−333. doi: 10.7506/spkx1002-6630-20230801-002
-
期刊类型引用(11)
1. 刘影,庞富,陈佳鸿,陈炯葵,蔡烁仪. 本草清咽润喉糖的配方优化及抗氧化研究. 农产品加工. 2024(21): 41-46 .  百度学术
百度学术
2. 张敏君,段雪伟,王燕,杨慧文,刘冰,向文静,由天辉. 构树根皮活性成分乙醇提取工艺优化及其抗氧化活性分析. 食品工业科技. 2023(11): 196-203 .  本站查看
本站查看
3. 王蕙雯. 豫西自然发酵柿子醋抗氧化性研究. 江苏调味副食品. 2023(03): 20-23 .  百度学术
百度学术
4. 裴文清,吕泸楠,王靖宇,浦思琦,雷霜,王春丽. 木瓜皮多酚和黄酮提取工艺优化及酪氨酸酶与胰脂肪酶抑制活性研究. 食品工业科技. 2022(01): 188-195 .  本站查看
本站查看
5. 周新崇,易灿,刘进兵. 微波辅助提取崀山脐橙皮总黄酮及生物活性研究. 邵阳学院学报(自然科学版). 2022(02): 87-95 .  百度学术
百度学术
6. 张清月,董姝慧,李胤豪,赵艳丽,史彬林,闫素梅. 诺丽果不同提取物抗氧化能力的比较研究. 中国粮油学报. 2022(05): 144-150 .  百度学术
百度学术
7. 关随霞,王蕙雯,杨肖瑞,郭淑敏,张翅,张培杰,李道敏. 大青叶总黄酮提取工艺优化及抗氧化性研究. 中国食品添加剂. 2022(09): 138-144 .  百度学术
百度学术
8. 陈慧玲,刘芳,钟恒勤,王伟枫. 超声波辅助乙醇提取百香果皮黄酮的工艺优化及黄酮抗氧化性测定. 宁德师范学院学报(自然科学版). 2022(03): 280-287 .  百度学术
百度学术
9. 任海云,韩瑞,张磊. 基于Box-Behnken响应面法优化党参抗氧化活性组分提取工艺. 中医药信息. 2022(12): 5-10 .  百度学术
百度学术
10. 赵雨晴,王宝庆,徐汉,刘楠楠. 醉鱼草总黄酮的提取及抗氧化活性研究. 化学试剂. 2021(07): 979-985 .  百度学术
百度学术
11. 杨青青,龚吉军. 响应面法优化超声辅助葛根浸提工艺及浸提液抗氧化活性研究. 食品安全质量检测学报. 2021(13): 5409-5417 .  百度学术
百度学术
其他类型引用(3)





 下载:
下载:

 下载:
下载:



