Research Progress of Molecular Biological Technology in Rapid Detection of Foodborne Pathogens
-
摘要: 食源性致病菌是影响食品安全的重要因素之一,高效快速的食源性致病菌检测技术对保障食品安全尤为重要。在食源性致病菌快速检测方法中,分子生物学技术因特异性强、精准度高、检测速度快等优势得到广泛应用。本文介绍了多种分子生物学技术在食源性致病菌快速检测中的应用,包括聚合酶链式反应(polymerase chain reaction,PCR)、环介导等温扩增(loop-mediated isothermal amplification,LAMP)、依赖核酸序列的扩增(nuclear acid sequence-based amplification,NAS-BA)、重组酶聚合酶扩增(recombinase polymerase amplification,RPA)、重组酶介导的等温核酸扩增(recombinase-aided isothermal amplification,RAA)、基因芯片(gene chip,GC),并对这些技术的应用现状、适用范围以及存在的问题等进行了分析总结,以期对未来的食源性致病菌快速检测技术提供理论参考。Abstract: Foodborne pathogens are one of the important factors affecting food safety. Efficient and rapid detection technology of foodborne pathogens is particularly important to ensure food safety. Among the rapid detection methods of foodborne pathogens, molecular biology technology has been widely used because of its advantages of strong specificity, high accuracy, and fast detection speed. This paper introduces the application of various molecular biological techniques in the rapid detection of foodborne pathogens, including polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), nuclear acid sequence-based amplification (NAS-BA), recombinase polymerase amplification (PRA), recombinase-aided isothermal amplification (RAA), and gene chip (GC). The application status, application scope and existing problems of these technologies are also analyzed and summarized to provide theoretical reference for the rapid detection of foodborne pathogens in the future.
-
Keywords:
- molecular biology technology /
- foodborne pathogens /
- rapid detection /
- food safety
-
俗话说“民以食为天,食以安为先”,因细菌引起的食源性疾病是我国重要的食品安全问题之一[1]。由食源性致病菌造成的食品安全问题时有发生,严重威胁着人民群众的身体健康,影响经济发展与社会稳定。尤其是随着现代生活节奏的加快及人们对健康和食品安全的关注度日益提高,消费者对最低限度加工的即食产品的需求不断增加,已开始引起公共卫生机构对食品安全保障的关注。因此对食品中的食源性致病菌进行准确检测具有重要意义[2]。
食源性致病菌检测一般可分为传统培养法检测和快速检测两种。传统培养法检测技术存在检测周期长、灵敏度低、操作繁琐等问题[3−4],无法满足及时得到检测结果的需要。灵敏高效的检测方法对于食品中的食源性病原体的快速检测,防止食源性疾病的暴发和食源性病原体的传播具有重要意义,尤其是近几十年来迅速发展的分子生物学快速检测技术通常比常规培养方法更灵敏、更特异、更省时、更省力、更可靠[5−8],因此得到广泛应用。
本文综合叙述了包括聚合酶链式反应(polymerase chain reaction,PCR)、多重PCR(multiplex PCR,mPCR)、实时荧光定量PCR(quantitative real-time PCR,qPCR)、数字PCR(digital PCR,dPCR)、环介导等温扩增(loop-mediated isothermal amplification,LAMP)、依赖核酸序列的扩增(nuclear acid sequence-based amplification,NAS-BA)、重组酶聚合酶扩增(recombinase polymerase amplification,RPA)、重组酶介导的等温核酸扩增(recombinase-aided isothermal amplification,RAA)、基因芯片(gene chip,GC)在食源性致病菌快速检测中的应用,并对分子生物学检测技术未来发展方向进行了展望,为分子生物学技术更好地应用于食源性致病菌快速检测提供参考。
1. 聚合酶链式反应
1.1 常规聚合酶链式反应
聚合酶链式反应(polymerase chain reaction,PCR)是一种特异性扩增目标DNA的技术,反应过程包括“变性、退火(复性)、延伸”三个阶段,其原理是目标双链DNA在热作用下变性解旋为单链DNA,然后基于DNA聚合酶的作用和碱基互补配对原则实现目标DNA的拷贝[9]。理论上来说,PCR体系中只要提供足够的酶与反应底物,重复“变性、退火、延伸”三个步骤即可在短时间内实现目标DNA的指数级扩增,借助凝胶电泳等方法观察扩增产物即可达到检测目的。
1992年,Rahn等[10]第一次将PCR技术应用于食源性致病菌检测,其根据沙门氏菌(Salmonella)的invA基因设计了PCR反应体系,对100种血清型的630株沙门氏菌进行了检测。除两株利赤非尔德沙门氏菌(S. litchfield)和两株山夫顿堡沙门氏菌(S. senftenberg)外,所有沙门氏菌均可检测到,检测率为99.4%。随着科学技术的发展,常规PCR检测技术已经非常成熟,是目前接受度最广泛的快速检测技术之一[11−12]。但常规PCR已经不能满足快速检测的需要,人们通过不断改良、与其他技术相结合,衍生出多种PCR技术[13−14]。
1.2 多重PCR
多重PCR(multiplex PCR,mPCR)又称多重引物PCR或复合PCR[15]。常规PCR只能扩增出一种DNA片段,即只能检测单一细菌,而mPCR通过在一个反应体系中针对某一种细菌的不同基因或多种细菌的不同基因设计多对特异性引物,通过优化反应条件,避免靶序列与不同引物间的交叉反应与相互干扰,可实现一次mPCR扩增多种DNA片段[16−17]。
1.2.1 单次mPCR检测一种细菌
在mPCR体系中添加单一菌种的基因组和对应的多对特异性引物,一次扩增出单一菌种的不同DNA片段。Feng等[18]根据4 h血清型单核细胞增生李斯特氏菌独有的基因LMxysn_1095、lmo1083和smcL设计了mPCR反应体系,成功区分出4 h L型菌株。谭燕等[19]根据鼠伤寒沙门氏菌的mgtC、inva、mogA、sseL、araB基因设计特异性引物,通过常规PCR测试了五种引物特异性,最终筛选出mgtC、mogA、araB基因设计了mPCR反应体系。经优化后,该体系的最低检测灵敏度为1.659×10−2 ng/μL。
相比于常规PCR,mPCR通过多个特异性特征位点对同一菌种进行鉴别扩增,提高了准确度,减少了假阳性结果的产生。
1.2.2 单次mPCR检测多种细菌
在mPCR体系中添加多个菌种的基因组和对应的多对特异性引物,可同时扩增出多个菌种的不同DNA片段[20]。翁思聪等[21]根据金黄色葡萄球菌的耐热核酸酶基因nuc、沙门氏菌的侵袭蛋白基因invA、志贺氏菌的侵染性质粒抗原H基因ipaH和副溶血性弧菌的毒力表达调控基因toxR设计了一种可快速检测水产品中4种食源性致病菌的mPCR反应体系。与国标方法对比,4种食源性致病菌整体符合率达94%以上,两种检测方法无显著性差异。
相比于常规PCR,mPCR通过一次反应可扩增出不同菌种的DNA片段,实现单次反应即可检测出多种目标细菌,因简洁、省时、高效、成本低等优点,具有更高的实用价值[22]。但在反应过程中由于多对引物及靶细菌基因组在同一体系中进行扩增,它们之间可能相互影响,因此mPCR在方法建立过程中需要精心优化反应体系,才能获得理想的扩增效果。
1.3 实时荧光定量PCR
实时荧光定量PCR(quantitative real-time PCR,qPCR)是在常规PCR的基础上发展出的第二代核酸定量检测技术[23],其原理是在PCR体系中加入荧光标记物,随着qPCR的进行,反应产物不断增加,荧光信号也不断增强,利用检测设备对荧光信号积累情况实时监测,最后通过荧光扩增曲线对扩增结果进行定量分析[24−25]。根据加入的荧光标记物,qPCR分为添加荧光染料的qPCR和添加荧光标记探针的qPCR两种。
1.3.1 添加荧光染料的qPCR
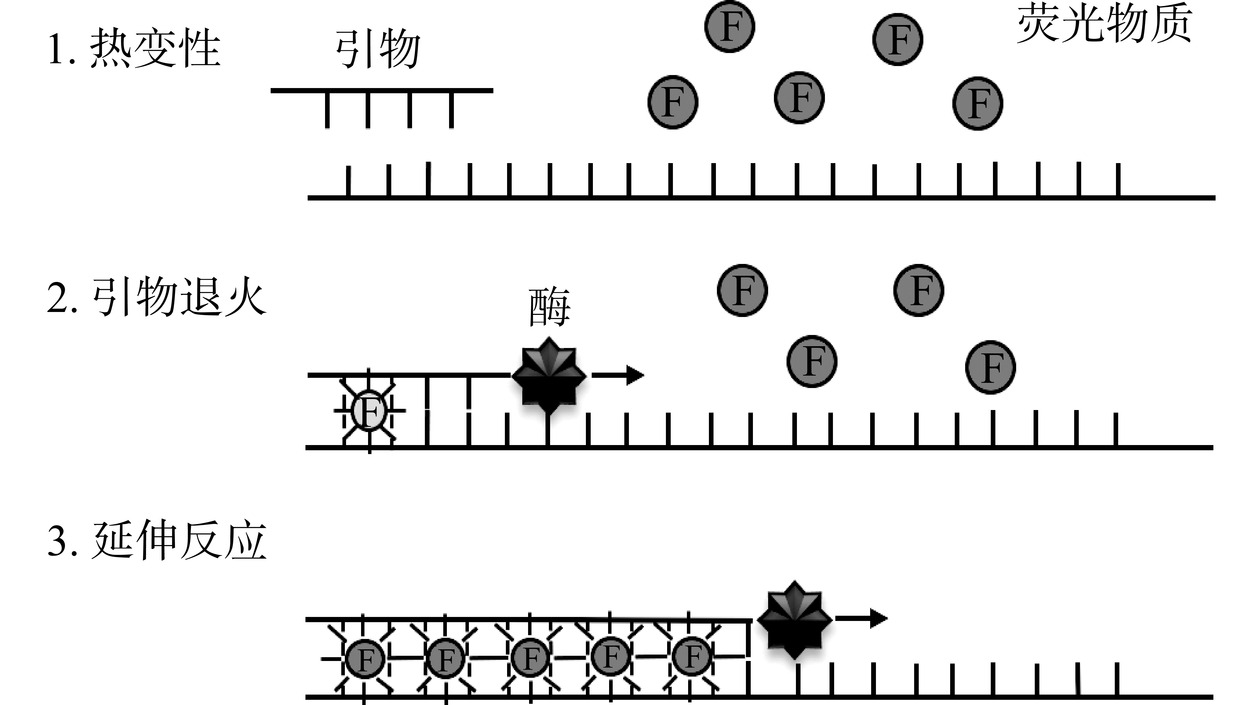
常用的荧光染料有SYBR Green I、YO-PRO、溴化乙锭、SYBR Gold等[26−28],其中SYBR Green I染料应用最广泛。在qPCR反应体系中加入过量的荧光染料,荧光染料结合双链DNA前不产生荧光或产生的荧光极弱;反应开始后,荧光染料非特异性掺入DNA双链,开始产生明显的荧光信号,根据监测荧光信号生成的扩增曲线可以实现结果的定量分析[29−31]。原理图如图1所示。Kumar等[32]利用SYBR Green I染料设计沙门氏菌qPCR检测方法用于快速定量检测海鲜中的沙门氏菌,大幅度缩短了检测时间。Fusco等[33]根据金黄色葡萄球菌肠毒素基因设计特异性引物,利用SYBR Green I染料建立了金黄色葡萄球菌qPCR检测方法,应用于牛奶的检测,灵敏度较常规PCR提高了3个数量级,检测准确率达96%。
添加荧光染料的qPCR成本低,使用简便,灵敏度高,但荧光染料与双链DNA为非特异性结合,对目标DNA无选择性,与添加荧光标记探针的qPCR相比特异性较弱,容易出现假阳性结果。
1.3.2 添加荧光标记探针的qPCR
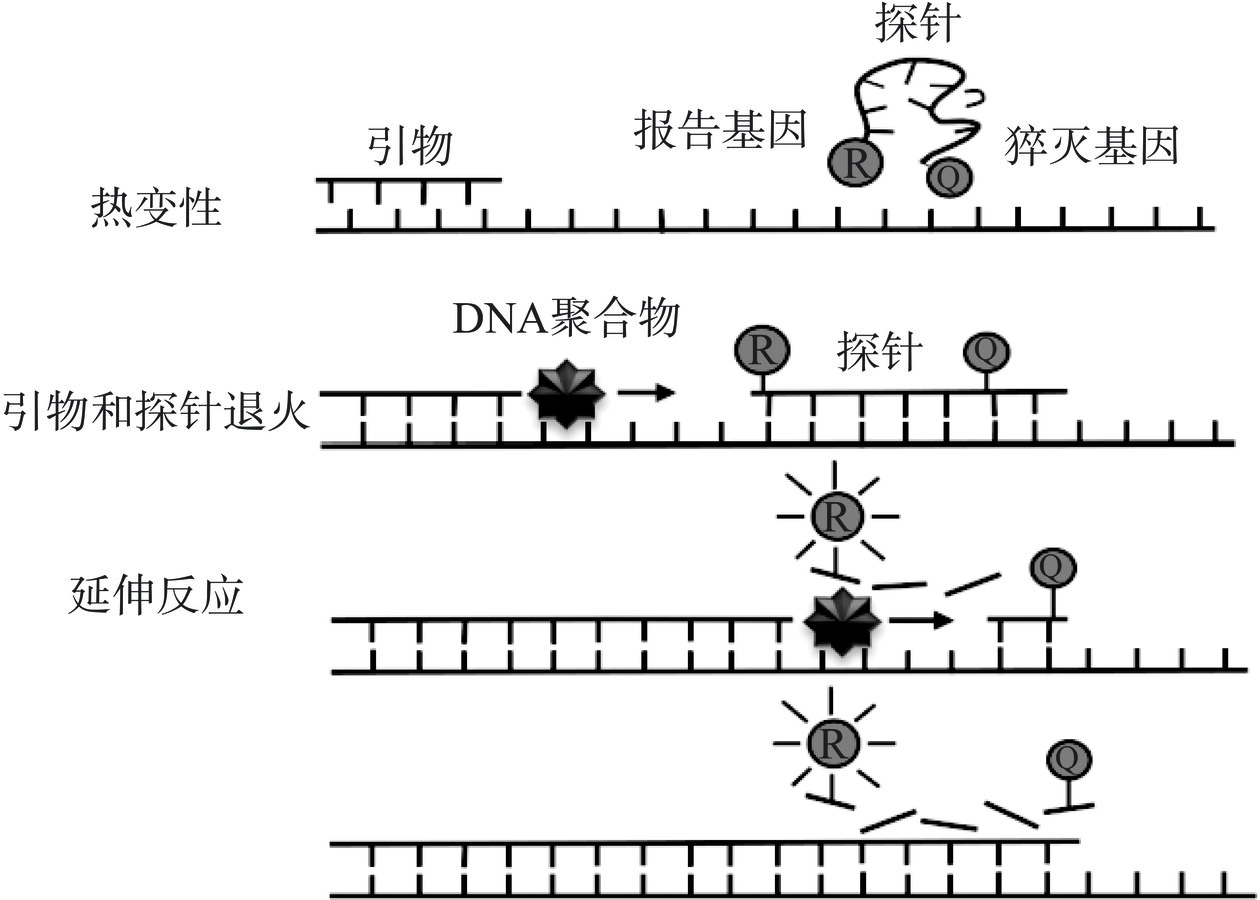
常用的荧光探针有TaqMan、TaqMan-MGB、Molecular beacon等[34−36],其中TaqMan探针应用最广泛。探针的两端分别为荧光报告基团和猝灭基团,完整的探针中的荧光标记基团发出的荧光信号会被猝灭基团吸收,不产生荧光。扩增开始前,完整的探针结合在目标DNA的任意一条单链上;扩增开始后,具有5’-3’外切酶活性的Taq DNA聚合酶将探针酶切降解,荧光标记基团和猝灭基团分离,释放出荧光标记基团,此时检测设备可接收到荧光信号。每扩增一个循环得到一条DNA双链,就会形成一个荧光分子,荧光信号强度也会同比例提高[37]。通过荧光信号强度的变化来监测扩增产物的变化量,从而得到相对应的荧光扩增曲线[38]。原理图如图2所示。王建昌等[39]根据志贺氏菌属高度保守的ipaH基因序列设计特异性引物和寡核苷酸探针,建立了检测志贺氏菌的qPCR方法。该方法在6 h内即可得到检测结果,最低检测限为9×103 CFU/mL。李丹丹等[40]以金黄色葡萄球菌Sa442基因为靶基因设计特异性引物和寡核苷酸探针,构建检测金黄色葡萄球菌的qPCR方法。该方法检测灵敏度为7.5 CFU/mL。利用该检测方法对采集的175份样品进行检测,共计检出5份阳性样品,与国标法(GB 4789.10)检测结果一致。
添加荧光标记探针的qPCR检测速度快,特异性高、准确性强,实现了快速检测技术从定性到定量的飞跃。但探针价格较高,而且需要专业的仪器设备和经验丰富的操作人员,限制了该技术的广泛应用,尤其是无法实现在现场快速检测中的应用。
1.3.3 多重荧光定量PCR
多重荧光定量PCR(multiplex quantitative real-time PCR,mqPCR)是将mPCR与qPCR相结合衍生出来的PCR技术。其原理是在同一qPCR反应体系中加入多对荧光标记物,实现一次mqPCR针对多种细菌进行定量分析。章沙沙[41]利用多种特异性引物和寡核苷酸探针设计了沙门氏菌、单核细胞增生李斯特氏菌、金黄色葡萄球菌、志贺氏菌、蜡样芽孢杆菌和副溶血性弧菌6种食源性致病菌的mqPCR检测方法,最低检测限可达到1×103 CFU/mL,在2 h内完成检测,大大缩短了检测时间。索原杰[42]基于金黄色葡萄球菌nuc基因、单核细胞增生李斯特氏菌hlyA基因及沙门氏菌orgC基因设计了多种特异性引物和寡核苷酸探针,构建了同时检测上述三种细菌的mqPCR方法,并在此基础上添加叠氮溴化丙锭(propidium monoazide,PMA)核酸染料,根据细胞膜对PMA的选择透过性(只能透过死细胞)、PMA与DNA发生共价交联反应,抑制死亡菌的DNA扩增,成功构建出PMA多重荧光定量活菌检测方法。
mqPCR的出现实现了一次qPCR反应定量检测多种食源性致病菌,极大提高了食源性致病菌快速检测的速度与准确性。但由于可获得的保守基因有限,多对特异性引物之间相互影响,实际操作中难度较高。
1.4 数字PCR
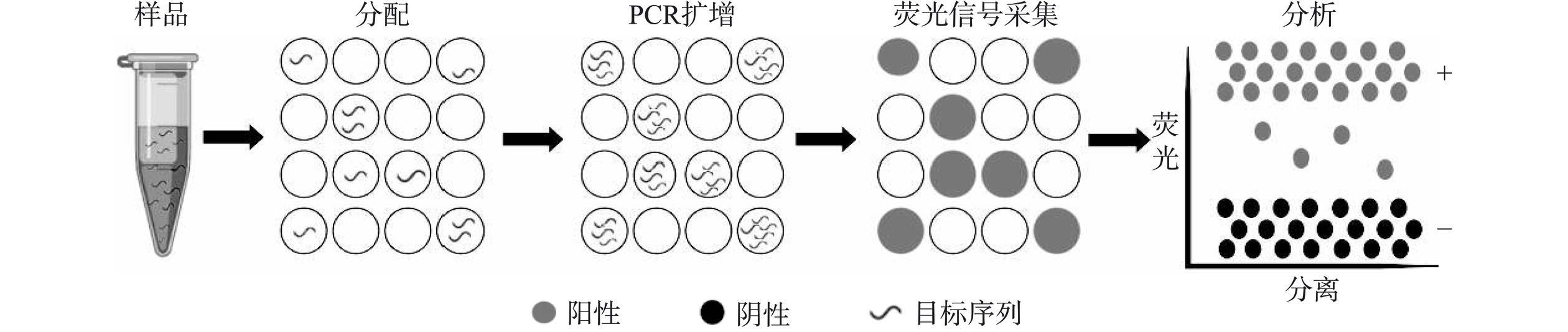
数字PCR(digital PCR,dPCR)是继第一代PCR(常规PCR)、第二代PCR(qPCR)之后的第三代PCR技术[43−45]。dPCR反应分为PCR扩增和荧光信号采集两个阶段[44]。PCR扩增阶段首先将参与反应的核酸样品分配到几万至几百万个反应单元中,使得每个反应单元含有一个或多个模版分子,之后对每一个反应单元进行PCR扩增。荧光信号采集阶段,读取每一个反应单元的荧光信号,对反应结果进行判断。最后,利用统计学方法,根据泊松分布原理和荧光信号阳性反应单元比例计算出目标核酸序列的拷贝数和浓度。原理图如图3所示。根据分液方式,dPCR划分为微流体式dPCR(microfluidic dPCR,mdPCR)[46]、微滴式dPCR(droplet dPCR,ddPCR)[47]和芯片式dPCR(chip dPCR,cdPCR)[48],其中最常用的为ddPCR。
程逸宇等[49]选取单核细胞增生李斯特氏菌iap为靶基因设计特异性引物及寡核苷酸探针,构建了快速检测乳产品中单核细胞增生李斯特氏菌的ddPCR方法。该方法能够对含菌量在5×103~5×106 CFU/mL之间的乳产品进行准确定量检测,检测下限和定量检测下限分别为5×102、5×103 CFU/mL。翁文川等[50]以副溶血性弧菌的tlh基因为目标基因设计筛选特异性引物及寡核苷酸探针,同时应用PMA核酸染料抑制死亡菌的DNA扩增,构建了快速检测水产品中副溶血性弧菌的PMA-ddPCR方法。PMA-ddPCR与PMA-qPCR的最低检出限分别为2×101、2×102 CFU/mL,PMA-ddPCR的检测灵敏度高于PMA-qPCR。应用PMA-ddPCR方法检测污染的基围虾和小帆立贝,两种海产品的最低检出限分别为1.9×101、8.9 CFU/g。
dPCR在操作方面更为简便,灵敏度高、重现性好、特异性强、不依赖标准曲线和Ct值,且能做到对目标序列的绝对定量,对特异性模板扩增抑制剂等干扰因素的耐受性更强,能够实现2~3 h内对低浓度的核酸样品进行精确定量[51]。dPCR虽具有上述优点以及巨大的应用潜力,但仍存在一定的局限性。由于dPCR容纳样品的体积小,导致动态范围较窄、稳定性和精密度略低于qPCR;dPCR数据分析平台众多,且处理方式不尽相同,因此对数据进行规范的协同分析也是dPCR今后发展的重点[52]。
2. 等温扩增技术
2.1 环介导等温扩增技术
环介导等温扩增(loop-mediated isothermal amplification,LAMP)技术通过对目标DNA的6个或8个特定区域设计2对或3对引物(一对外部引物、一对内部引物,有时增加一对环引物)进行特异性识别,利用Bst链置换DNA聚合酶在60~65 ℃等温条件扩增,其步骤包括“模板合成、循环扩增、延长再循环”三个阶段,整个反应持续约1 h[53−55]。黄朱梁等[56]根据单增李斯特氏菌hlyA基因设计了2对特异性引物,建立了LAMP快速检测技术,在60 min内即可得到检测结果,并利用实时浊度计和琼脂糖凝胶电泳相互验证,二者结果一致。关玉婷等[57]根据沙门氏菌stn基因设计了2对特异性引物,应用羟基萘酚蓝(hydroxy naphthol blue,HNB)在LAMP反应中的颜色变化特征,建立了LAMP-HNB可视化检测方法,仅需30 min即可得到准确结果,且对3株沙门氏菌和8株非沙门氏菌表现出良好的特异性。纪懿芳等[58−59]以副溶血性弧菌tlh基因设计特异性引物,建立的副溶血性弧菌的LAMP快速检测技术,最低检出限为2.8×102 CFU/mL。该团队将LAMP技术与HNB结合建立的LAMP-HNB可视化检测技术,可根据反应体系颜色变化直接判断反应结果,与常规PCR检测结果一致,无显著差异。
LAMP技术产物检测和结果观察主要有四种方法:LAMP技术诞生初期,研究人员使用凝胶电泳方法对其扩增产物进行检测,根据产生的条带判断结果[60]。但LAMP扩增产物电泳呈阶梯状,难以判断结果,而且极易形成气溶胶污染[61−62]。后来,由于观察到LAMP技术在扩增过程中会生成焦磷酸镁沉淀,反应液的浊度会发生变化,因此进行了改进,通过测定反应体系浊度变化实现可视化检测,但通过浊度进行判断,误差较大[63−64]。进一步研究发现,在LAMP反应体系中加入荧光染料,利用荧光透色仪测定反应体系的实时荧光值可实现LAMP技术可视化实时定量检测,同时避免了气溶胶污染[65−66]。最新的LAMP技术应用HNB,使阳性结果直接呈现深蓝色,阴性结果呈现为绿色[67−68],更易于观察。
LAMP是一种创新的基因扩增技术,具有操作简单、无需复杂昂贵的设备、不需要进行热循环等优点,且经过多年的发展,易受污染、结果可能出现假阳性等问题也得到了改善。目前,LAMP已成为一种成熟可靠的分子生物学检测方法,在监测和控制食源性疾病的传播方面发挥了重要作用。
2.2 依赖核酸序列的扩增技术
依赖核酸序列的扩增(nuclear acid sequence-based amplification,NAS-BA)技术是一种以RNA为模板,依靠逆转录酶、RNA酶H、T7 RNA聚合酶与两个特异性引物进行反应的恒温快速扩增技术[69]。第一个特异性引物与目标RNA结合后,在逆转录酶的作用下合成互补DNA单链;在RNA酶H作用下,目标RNA发生降解,保留互补DNA单链;第二个特异性引物与互补DNA单链结合,在T7 RNA聚合酶作用下,大量合成目标RNA[70−71]。通过琼脂糖凝胶电泳观察结果[72]。Zhai等[73]利用mRNA逆转录设计靶向沙门氏菌特异性xcd基因的NAS-BA体系,成功检测出猪肉、牛肉和牛奶中的沙门氏菌,最低检出限为10 CFU/25 g。倪鑫等[74]根据副溶血性弧菌的tlh基因为靶基因设计特异性引物,建立了快速检测副溶血性弧菌的NAS-BA技术。该NAS-BA方法对副溶血性弧茵的最小检出量为5.1×102 CFU/mL,高于常规PCR技术,而且与该方法建立过程中所用非特异性种属的菌无任何交叉反应。
NAS-BA技术操作简单,整个反应在恒温条件下进行,不需要特殊的控温设备。但NAS-BA技术扩增产物为RNA,极易发生降解,且在染色能力上RNA不如DNA高效,容易出现假阳性的问题[75],这些缺点一定程度上限制了NAS-BA技术的推广。
2.3 重组酶聚合酶扩增技术
重组酶聚合酶扩增(recombinase polymerase amplification,RPA)技术是继LAMP技术之后的另一种恒温快速扩增技术[76]。RPA技术通过模拟T4噬菌体内的核酸复制,于体外恒温环境中,在重组酶的作用下实现与目标DNA的特异性识别与结合,在单链DNA结合蛋白的作用下特异性解旋目标DNA的双螺旋结构,在链置换DNA聚合酶的作用下实现目标DNA的扩增[77−78]。洪晗露[79]根据阪崎克罗诺杆菌ompA基因、沙门氏菌invA基因设计特异性探针与引物建立了双重PRA反应体系,对阪崎克罗诺杆菌的最低检测限为1×104 CFU/mL,对沙门氏菌的最低检测限为1×103 CFU/mL,无交叉反应。刘立兵等[80−81]利用沙门氏菌invA基因的保守序列设计特异性引物,建立了沙门氏菌RPA快速检测技术,对反应条件进行优化后,在20 min内即可完成对目标DNA的扩增。之后该团队根据志贺氏菌ipaH基因的保守序列设计引物及exo探针,将荧光技术与RPA技术结合,建立了快速检测志贺氏菌的实时荧光RPA技术。
RPA技术对实验环境要求低,整个反应不需要高温变性,在37~42 ℃下可以完成扩增,反应时间在5~45 min,耗时较短[82]。RPA技术的特异性引物设计较为简单,仅需要一对特异性引物即可达到对目标DNA特异性的扩增。RPA技术的设备要求低、检测所需时间短、灵敏度高、特异性好使其成为现场快速检测的首选[83],但在检测过程中应注意气溶胶的产生与扩散,防止交叉污染[84]。
2.4 重组酶介导的等温核酸扩增技术
重组酶介导的等温核酸扩增(recombinase aided amplification,RAA)技术同样是继LAMP技术之后的另一种快速恒温核酸扩增技术[85]。与PRA技术的区别在于PRA技术中的重组酶来源于T4噬菌体,而RAA技术中的重组酶来自于细菌或真菌[86−87]。RAA技术的重组酶与引物紧密结合,形成酶-引物聚合体,特异性识别并结合目标DNA,在单链DNA结合蛋白作用下,将目标DNA内部双链结构展开的同时使引物和模板配对,随后在链置换DNA聚合酶作用下,组成新的双链目标DNA,不断重复此过程即实现目标DNA的扩增[87−88]。
郝林慧等[89]根据副溶血性弧菌tlh基因序列设计特异性引物,经引物筛选,建立了快速检测副溶血性弧菌RAA技术,在30 min内即可观察到检测结果。该方法与其他弧菌属细菌无交叉反应,检出限为5.3 CFU/mL,且RAA技术和传统培养法结果符合率为100%,表现出高度一致性。黄新新等[90]根据单核细胞增生李斯特氏菌hlyA基因设计了特异性引物和探针,建立了快速检测单核细胞增生李斯特氏菌荧光RAA技术,20 min即可完成扩增,5 min之内即可获得检测结果,检出限为3×102 CFU/mL。
RAA技术与RPA技术优缺点较为相似,RAA技术目前主要应用于检测食品中的寄生虫,对食源性致病菌应用较少,拥有广阔的发展前景。
3. 基因芯片
基因芯片(gene chip,GC)又称DNA微阵列(DNA microarray)、DNA芯片[91−93],利用固相原位合成、显微点样等微加工技术,将数以万计的已知序列的特定DNA片段(DNA探针)有规律地固定在玻璃芯片、硅芯片等支持物上,构成一个二维DNA探针序列,与标记后的待测样品按照碱基互补配对原则进行杂交,通过放射自显影技术、激光共聚焦荧光检测技术等扫描芯片,对杂交信号进行分析检测,最终获得样品的DNA序列[94]。Day等[95]基于流式荧光(Luminex xMAP)技术设计Bio-Plex悬浮芯片,并通过顺磁性微球-抗体复合物特异性检测单核细胞增生李斯特氏菌分泌蛋白和膜结合蛋白。对哈密瓜和包装沙拉样品进行检测表明,单核细胞增生李斯特氏菌的最低检测限为100 CFU/g。Hu等[96]根据鼠伤寒沙门氏菌、副溶血性弧菌、金黄色葡萄球菌、单核细胞增生李斯特氏菌和大肠杆菌O157:H7的特异性序列,从3227个杂交探针中筛选出141个特异性探针(鼠伤寒沙门氏菌20个、副溶血性弧菌46个、金黄色葡萄球菌24个、单核细胞增生李斯特氏菌26个、大肠杆菌O157:H7 25个)。通过原位合成技术将141个特异性探针序列平铺在微流控芯片上,对鲜切哈密瓜和莴苣中的五种目标食源性致病菌进行检测,最低检测限为3 log CFU/g。
GC是随着人类基因组计划(human genome project)的逐步实施以及分子生物学相关学科的迅猛发展而诞生的一种新型快速检测技术,与其他快速检测技术相比,GC可以一次性对含有多种序列的样品进行检测和分析,解决了传统核酸检测技术步骤繁杂、操作序列数量少、自动化程度低、检测效率低等问题[97]。但GC同样也存在制作繁琐、检测成本过高、前期需要投入大量的人力物力、缺少标准化程序、需要专业人员进行操作等[98]问题,因此GC的简易制作、标准化操作成为该技术日后的主要发展与改进方向。
4. 总结
上述各种食源性致病菌分子生物学快速检测技术对温度要求和引物数量各不相同,PCR反应过程中通常伴随着温度的变化。常规PCR通过1对引物便可进行检测,但是对于温度条件要求较高,耗时长且容易受到污染。为了实现实验需求,通过增加引物数量、添加燃料和改变反应模板等方法对常规PCR进行改善能够实现扩大检测数量、提高检测的灵敏度和缩短反应时间等目的,但同时也会增加实验成本和难度,影响实验的稳定性。其他的分子检测技术通常是在恒温条件下进行的,不同的反应所需的条件以及难易程度也不相同,所呈现的结果各有优势,需针对具体研究选择最适合的检测方法。具体方法、反应条件、引物类型以及优势和不足如表1所示。
表 1 不同食源性致病菌分子生物学快速检测方法对比Table 1. Comparison of rapid molecular detection methods of different foodborne pathogens方法 温度 引物数量 产物类型 特点 不足 常规PCR 变温 1对 DNA 引物设计简单、特异性强、应用广泛 温度要求高、易受污染、耗时较长 mPCR 变温 多对 DNA 可同时检测多种食源性致病菌、特异性强 引物设计复杂、扩增效果不佳 qPCR 变温 1或多对 DNA 灵敏度高、检测速度快、可实时观测 价格昂贵、引物设计复杂、需要专业设备和人员 dPCR 变温 1对 DNA 灵敏度高、特异性强、重现性强、耐受性强、绝对定量分析 准确性略差、稳定性略差、数据分析平台不统一 LAMP 37 ℃恒温 2或3对 DNA 扩增效率高、特异性强、可视化检测、设备要求低 引物设计复杂、易产生气溶胶污染、易产生假阳性 NAS-BA 37 ℃恒温 2对 RNA 灵敏度高、特异性强、检测速度快、不易受污染 需要逆转录酶、样品制备复杂、产物容易分解 RPA 37 ℃恒温 1对 DNA 特异性强、灵敏度高、恒温检测 试剂昂贵、引物较长,不适用于短序列的核酸检测 RAA 37 ℃恒温 1对 DNA 特异性强、灵敏度高、恒温检测 试剂昂贵、引物较长,不适用于短序列的核酸检测 GC / / / 高通量检测、自动化程度高、步骤精简、灵敏度高 制作过程繁琐、检测成本高、缺少标准化程序 5. 展望
食品是人类赖以生存的物质基础,食品安全是人民和国家乃至国际社会持续关注的重点问题之一,对食源性致病菌快速检测是保障食品安全的一道重要防线。随着DNA及基因组测序技术不断成熟,各种致病菌基因信息的获得将更容易、更迅速,以之为基础的分子生物学技术在食源性致病菌快速检测中的作用也将越来越重要。
分子生物学快速检测技术发展到现在已基本能够满足食品快速检测中快速、高特异性、高灵敏度等需要,且有能自动化、可多重检测、结果可靠等优点,但它们并不是完美的,仍旧存在价格昂贵、引物设计复杂、扩增产物一般难以定量、反应体系易受污染、不能检测毒素、一般不能区分血清型、需要纯化DNA、易受抑制剂的影响、不能用于现场检测和实时检测等问题。因此,针对食源性致病菌的分子生物学快速检测技术未来主要的发展方向主要有以下几个方面:a.与其他新型技术结合,如与免疫学技术、光学技术、电化学技术、生物传感器技术等结合,多种技术取长补短,开发出更为廉价、快速、便携的检测技术;b.进一步优化设计引物程序,降低设计引物的难度,避免不同引物交叉反应与相互干扰,避免假阳性;c.在现有技术上不断完善,研究如何在现有技术上对食源性致病菌进行定量检测。
随着科学技术的不断进步,检测仪器将会向便携化、简易化、数字化、智能化方向发展。多种检测技术联用、更高灵敏度、更高准确性、更强特异性、更广适用范围、更快检测速度是未来食源性致病菌检测技术的发展趋势。
-
表 1 不同食源性致病菌分子生物学快速检测方法对比
Table 1 Comparison of rapid molecular detection methods of different foodborne pathogens
方法 温度 引物数量 产物类型 特点 不足 常规PCR 变温 1对 DNA 引物设计简单、特异性强、应用广泛 温度要求高、易受污染、耗时较长 mPCR 变温 多对 DNA 可同时检测多种食源性致病菌、特异性强 引物设计复杂、扩增效果不佳 qPCR 变温 1或多对 DNA 灵敏度高、检测速度快、可实时观测 价格昂贵、引物设计复杂、需要专业设备和人员 dPCR 变温 1对 DNA 灵敏度高、特异性强、重现性强、耐受性强、绝对定量分析 准确性略差、稳定性略差、数据分析平台不统一 LAMP 37 ℃恒温 2或3对 DNA 扩增效率高、特异性强、可视化检测、设备要求低 引物设计复杂、易产生气溶胶污染、易产生假阳性 NAS-BA 37 ℃恒温 2对 RNA 灵敏度高、特异性强、检测速度快、不易受污染 需要逆转录酶、样品制备复杂、产物容易分解 RPA 37 ℃恒温 1对 DNA 特异性强、灵敏度高、恒温检测 试剂昂贵、引物较长,不适用于短序列的核酸检测 RAA 37 ℃恒温 1对 DNA 特异性强、灵敏度高、恒温检测 试剂昂贵、引物较长,不适用于短序列的核酸检测 GC / / / 高通量检测、自动化程度高、步骤精简、灵敏度高 制作过程繁琐、检测成本高、缺少标准化程序 -
[1] 刘耀, 魏元苗, 李玲, 等. 食源性致病菌溯源分型技术研究进展[J]. 食品工业科技,2022,43(12):427−437. [LIU Yao, WEI Yuanmiao, LI Ling, et al. Advances in traceability typing and identification of foodborne pathogens[J]. Science and Technology of Food Industry,2022,43(12):427−437.] LIU Yao, WEI Yuanmiao, LI Ling, et al. Advances in traceability typing and identification of foodborne pathogens[J]. Science and Technology of Food Industry, 2022, 43(12): 427−437.
[2] 董晶, 卢鑫, 郭威, 等. 等温扩增技术在食源性致病菌检测中的研究进展[J]. 食品与发酵工业,2021,47(8):256−260. [DONG Jing, LU Xin, GUO Wei, et al. Research progress on isothermal amplification technology in the detection of foodborne pathogens[J]. Food and Fermentation Industries,2021,47(8):256−260.] DONG Jing, LU Xin, GUO Wei, et al. Research progress on isothermal amplification technology in the detection of foodborne pathogens[J]. Food and Fermentation Industries, 2021, 47(8): 256−260.
[3] 王纯, 张若鸿, 王晓芳, 等. 食源性致病菌活菌检测方法的应用现状及展望[J]. 食品研究与开发,2021,42(23):183−189. [WANG Chun, ZHANG Ruohong, WANG Xiaofang, et al. Application status and prospects of detection methods for viable foodborne pathogens[J]. Food Research and Development,2021,42(23):183−189.] doi: 10.12161/j.issn.1005-6521.2021.23.029 WANG Chun, ZHANG Ruohong, WANG Xiaofang, et al. Application status and prospects of detection methods for viable foodborne pathogens[J]. Food Research and Development, 2021, 42(23): 183−189. doi: 10.12161/j.issn.1005-6521.2021.23.029
[4] 张偲偲, 刘兴泉, 杨媚婷, 等. 食源性致病菌现场即时检测技术研究进展[J]. 分析化学,2021,49(10):1631−1639. [ZHANG Sisi, LIU Xingquan, YANG Meiting, et al. Recent advances in point-of-care diagnosis for foodborne pathogens[J]. Chinese Journal of Analytical Chemistry,2021,49(10):1631−1639.] ZHANG Sisi, LIU Xingquan, YANG Meiting, et al. Recent advances in point-of-care diagnosis for foodborne pathogens[J]. Chinese Journal of Analytical Chemistry, 2021, 49(10): 1631−1639.
[5] 钟宜科, 邹大阳, 赵彤, 等. 食源性致病菌核酸检测技术研究进展[J]. 食品研究与开发,2020,41(12):218−224. [ZHONG Yike, ZOU Dayang, ZHAO Tong, et al. Advances in nucleic acid-based methods for detecting foodborne pathogen[J]. Food Research and Development,2020,41(12):218−224.] doi: 10.12161/j.issn.1005-6521.2020.12.036 ZHONG Yike, ZOU Dayang, ZHAO Tong, et al. Advances in nucleic acid-based methods for detecting foodborne pathogen[J]. Food Research and Development, 2020, 41(12): 218−224. doi: 10.12161/j.issn.1005-6521.2020.12.036
[6] 王丹丹, 刘鸣畅, 杨艳歌, 等. 食源性致病菌快速检测技术研究进展[J]. 食品科学,2022,43(3):276−285. [WANG Dandan, LIU Mingchang, YANG Yange, et al. Recent progress in technologies for rapid detection of foodborne pathogens[J]. Food Science,2022,43(3):276−285.] doi: 10.7506/spkx1002-6630-20201105-048 WANG Dandan, LIU Mingchang, YANG Yange, et al. Recent progress in technologies for rapid detection of foodborne pathogens[J]. Food Science, 2022, 43(3): 276−285. doi: 10.7506/spkx1002-6630-20201105-048
[7] CAPOBIANCO J A, CLARK M, CARIOU A, et al. Detection of Shiga toxin-producing Escherichia coli (STEC) in beef products using droplet digital PCR[J]. International Journal of Food Microbiology,2020,319:108499. doi: 10.1016/j.ijfoodmicro.2019.108499
[8] 钟海霞, 周寒嫣, 罗欢, 等. 等温扩增技术在食源性致病菌检测中的研究进展[J]. 食品工业科技,2019,40(7):362−367. [ZHONG Haixia, ZHOU Hanyan, LUO Huan, et al. Research progress of isothermal amplification in the detection of pathogenic bacteria in food[J]. Science and Technology of Food Industry,2019,40(7):362−367.] ZHONG Haixia, ZHOU Hanyan, LUO Huan, et al. Research progress of isothermal amplification in the detection of pathogenic bacteria in food[J]. Science and Technology of Food Industry, 2019, 40(7): 362−367.
[9] UMESHA S, MANUKUMAR H M. Advanced molecular diagnostic techniques for detection of food-borne pathogens:Current applications and future challenges[J]. Critical Reviews in Food Science and Nutrition,2018,58(1):84−104. doi: 10.1080/10408398.2015.1126701
[10] RAHN K, DE GRANDIS S A, CLARKE R C, et al. Amplification of an invA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella[J]. Molecular and Cellular Probes,1992,6(4):271−279. doi: 10.1016/0890-8508(92)90002-F
[11] 孙颖颖, 董鹏程, 朱立贤, 等. 食源性致病菌快速检测研究进展[J]. 食品与发酵工业,2020,46(17):264−270. [SUN Yingying, DONG Pengcheng, ZHU Lixian, et al. Research progress in rapid detection of foodborne pathogens[J]. Food and Fermentation Industries,2020,46(17):264−270.] SUN Yingying, DONG Pengcheng, ZHU Lixian, et al. Research progress in rapid detection of foodborne pathogens[J]. Food and Fermentation Industries, 2020, 46(17): 264−270.
[12] 白亚龙, 索玉娟, 周昌艳. 食源性致病菌PCR检测前处理方法研究进展[J]. 食品与机械,2017,33(12):191−196. [BAI Yalong, SUO Yujuan, ZHOU Changyan. A review of the development in pretreatment methods for PCR detection of food-borne pathogens[J]. Food and Machinery,2017,33(12):191−196.] BAI Yalong, SUO Yujuan, ZHOU Changyan. A review of the development in pretreatment methods for PCR detection of food-borne pathogens[J]. Food and Machinery, 2017, 33(12): 191−196.
[13] 丁博群, 刘珊娜. 荧光定量PCR技术在食品快速检测中的应用[J]. 食品工业科技,2021,42(7):366−373. [DING Boqun, LIU Shanna. Application of real-time quantitative PCR technology in food rapid detection[J]. Science and Technology of Food Industry,2021,42(7):366−373.] DING Boqun, LIU Shanna. Application of real-time quantitative PCR technology in food rapid detection[J]. Science and Technology of Food Industry, 2021, 42(7): 366−373.
[14] 张明娟, 王娟, 袁磊, 等. 多重聚合酶链式反应技术在食源性致病菌检测上的应用研究进展[J]. 食品与发酵工业,2021,47(2):305−310. [ZHANG Mingjuan, WANG Juan, YUAN Lei, et al. Application of multiplex polymerase chain reaction in detection of foodborne pathogens[J]. Food and Fermentation Industries,2021,47(2):305−310.] ZHANG Mingjuan, WANG Juan, YUAN Lei, et al. Application of multiplex polymerase chain reaction in detection of foodborne pathogens[J]. Food and Fermentation Industries, 2021, 47(2): 305−310.
[15] ZHAO X, LIN C W, WANG J, et al. Advances in rapid detection methods for foodborne pathogens[J]. Journal of Microbiology and Biotechnology,2014,24(3):297−312. doi: 10.4014/jmb.1310.10013
[16] ZHANG Y, HU X, WANG Q. Sensitive and specific detection of E. coli, Listeria monocytogenes, and Salmonella enterica serovar Typhimurium in milk by microchip electrophoresis combined with multiplex PCR amplification[J]. Microchemical Journal,2020,157:104876. doi: 10.1016/j.microc.2020.104876
[17] ALÍA A, ANDRADE M J, CÓRDOBA J J, et al. Development of a multiplex real-time PCR to differentiate the four major Listeria monocytogenes serotypes in isolates from meat processing plants[J]. Food Microbiology,2020,87:103367. doi: 10.1016/j.fm.2019.103367
[18] FENG Y, YAO H, CHEN S, et al. Rapid detection of hypervirulent serovar 4 h Listeria monocytogenes by multiplex PCR[J]. Frontiers in Microbiology,2020,11:1309. doi: 10.3389/fmicb.2020.01309
[19] 谭燕, 肖紫鸣, 李小林, 等. 多重PCR技术检测食品中鼠伤寒沙门氏菌3种毒力基因[J]. 食品研究与开发,2021,42(12):171−176. [TAN Yan, XIAO Ziming, LI Xiaolin, et al. Detection of three virulence genes of Salmonella typhimurium in food by multiplex PCR[J]. Food Research and Development,2021,42(12):171−176.] doi: 10.12161/j.issn.1005-6521.2021.12.027 TAN Yan, XIAO Ziming, LI Xiaolin, et al. Detection of three virulence genes of Salmonella typhimurium in food by multiplex PCR[J]. Food Research and Development, 2021, 42(12): 171−176. doi: 10.12161/j.issn.1005-6521.2021.12.027
[20] MAO Y, HUANG X, XIONG S, et al. Large-volume immunomagnetic separation combined with multiplex PCR assay for simultaneous detection of Listeria monocytogenes and Listeria ivanovii in lettuce[J]. Food Control,2016,59:601−608. doi: 10.1016/j.foodcont.2015.06.048
[21] 翁思聪, 朱军莉, 励建荣. 水产品中4种常见致病菌多重PCR检测方法的建立及评价[J]. 水产学报,2011,35(2):305−314. [WENG Sicong, ZHU Junli, LI Jianrong. Establishment and evaluation on a multiplex-PCR method for detection of four pathogenic bacteria in aquatic products[J]. Journal of Fisheries of China,2011,35(2):305−314.] WENG Sicong, ZHU Junli, LI Jianrong. Establishment and evaluation on a multiplex-PCR method for detection of four pathogenic bacteria in aquatic products[J]. Journal of Fisheries of China, 2011, 35(2): 305−314.
[22] XU Y G, SUN L M, WANG Y S, et al. Simultaneous detection of Vibrio cholerae, Vibrio alginolyticus, Vibrio parahaemolyticus and Vibrio vulnificus in seafood using dual priming oligonucleotide (DPO) system-based multiplex PCR assay[J]. Food Control,2017,71:64−70. doi: 10.1016/j.foodcont.2016.06.024
[23] BARKALLAH M, GHARBI Y, SLIMA A B, et al. Simultaneous detection of Waddlia chondrophila and Listeria monocytogenes in aborted ruminant samples by real-time quantitative PCR[J]. Journal of Microbiological Methods,2016,125:64−69. doi: 10.1016/j.mimet.2016.04.001
[24] 朱耀磊, 桑雪, 甘未祺, 等. H-NS基因作为靶基因的荧光定量PCR快速检测海产品中的副溶血性弧菌[J]. 食品工业科技,2015,36(19):155−158,162. [ZHU Yaolei, SANG Xue, GAN Weiqi, et al. Rapid detection of Vibrio parahaemolyticus in seafood by real-time PCR targeted to the H-NS gene[J]. Science and Technology of Food Industry,2015,36(19):155−158,162.] ZHU Yaolei, SANG Xue, GAN Weiqi, et al. Rapid detection of Vibrio parahaemolyticus in seafood by real-time PCR targeted to the H-NS gene[J]. Science and Technology of Food Industry, 2015, 36(19): 155−158,162.
[25] WEI S, PARK B J, KIM S H, et al. Detection of Listeria monocytogenes using Dynabeads® anti-Listeria combined with real-time PCR in soybean sprouts[J]. LWT,2019,99:533−539. doi: 10.1016/j.lwt.2018.10.023
[26] 李月华, 周巍, 杨岚, 等. 葡糖杆菌的SYBR Green Ι荧光定量PCR方法的建立[J]. 现代食品科技,2015,31(4):272−276, 241. [LI Yuehua, ZHOU Wei, YANG Lan, et al. Establishment of real-time fluorescence quantitative PCR for Gluconobacter using SYBR Green I[J]. Modern Food Science and Technology,2015,31(4):272−276, 241.] LI Yuehua, ZHOU Wei, YANG Lan, et al. Establishment of real-time fluorescence quantitative PCR for Gluconobacter using SYBR Green I[J]. Modern Food Science and Technology, 2015, 31(4): 272−276, 241.
[27] 吴海江, 孙玉萍, 范田丽, 等. EMA结合实时荧光PCR方法检测单核细胞增生李斯特氏菌[J]. 食品与生物技术学报,2020,39(11):65−70. [WU Haijiang, SUN Yuping, FAN Tianli, et al. Detection of live Listeria monocytogenes by EMA-qPCR[J]. Journal of Food Science and Biotechnology,2020,39(11):65−70.] doi: 10.3969/j.issn.1673-1689.2020.11.009 WU Haijiang, SUN Yuping, FAN Tianli, et al. Detection of live Listeria monocytogenes by EMA-qPCR[J]. Journal of Food Science and Biotechnology, 2020, 39(11): 65−70. doi: 10.3969/j.issn.1673-1689.2020.11.009
[28] EINEN J, THORSETH I H, ØVREÅS L. Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy[J]. FEMS Microbiology Letters,2008,282(2):182−187. doi: 10.1111/j.1574-6968.2008.01119.x
[29] LAW J W F, AB MUTALIB N S, CHAN K G, et al. Rapid methods for the detection of foodborne bacterial pathogens:Principles, applications, advantages and limitations[J]. Frontiers in Microbiology,2015,5:770.
[30] KAWASE J, ETOH Y, IKEDA T, et al. Improved multiplex real-time SYBR Green PCR assay for analysis of 24 target genes from 16 bacterial species in fecal DNA samples from patients with foodborne illnesses[J]. Japanese Journal of Infectious Diseases,2016,69(3):191−201. doi: 10.7883/yoken.JJID.2015.027
[31] 李金峰. 三种食源性致病菌的多重实时PCR检测研究[D]. 武汉:华中农业大学, 2008. [LI Jinfeng. Establishment of triplex real-time PCR method for detection of foodborne pathogens[D]. Wuhan:Huazhong Agricultural University, 2008.] LI Jinfeng. Establishment of triplex real-time PCR method for detection of foodborne pathogens[D]. Wuhan: Huazhong Agricultural University, 2008.
[32] KUMAR R, SURENDRAN P K, THAMPURAN N. Rapid quantification of Salmonella in seafood using real-time PCR assay[J]. Journal of Microbiology and Biotechnology,2010,20(3):569−573.
[33] FUSCO V, QUERO G M, MOREA M, et al. Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc) and quantitative detection in raw milk by real time PCR[J]. International Journal of Food Microbiology,2011,144(3):528−537. doi: 10.1016/j.ijfoodmicro.2010.11.016
[34] 胡朝友. TaqMan探针荧光PCR法定量检测水中大肠埃希氏菌方法建立和应用[D]. 苏州:苏州大学, 2014. [HU Zhaoyou. Development of a real-time qPCR method (taqman probe based) for detection and enumeration of Escherichia coli in water[D]. Suzhou:Soochow University, 2014.] HU Zhaoyou. Development of a real-time qPCR method (taqman probe based) for detection and enumeration of Escherichia coli in water[D]. Suzhou: Soochow University, 2014.
[35] MCCLEARY S, HENSHILWOOD K. Novel quantitative TaqMan® MGB real-time PCR for sensitive detection of Vibrio aestuarianus in Crassostrea gigas[J]. Diseases of Aquatic Organisms,2015,114(3):239−248. doi: 10.3354/dao02869
[36] RANI N, VAJPAYEE P, BHATTI S, et al. Quantification of Salmonella typhi in water and sediments by molecular-beacon based qPCR[J]. Ecotoxicology and Environmental Safety,2014,108:58−64. doi: 10.1016/j.ecoenv.2014.06.033
[37] 邵美丽, 董鑫, 赵燕丽, 等. 单增李斯特菌和金黄色葡萄球菌双重荧光定量PCR检测方法建立[J]. 食品科学,2013,34(16):169−172. [SHAO Meili, DONG Xin, ZHAO Yanli, et al. A duplex fluorescence quantitative PCR assay for detecting Listeria monocytogenes and Staphylococcus aureus[J]. Food Science,2013,34(16):169−172.] doi: 10.7506/spkx1002-6630-201316034 SHAO Meili, DONG Xin, ZHAO Yanli, et al. A duplex fluorescence quantitative PCR assay for detecting Listeria monocytogenes and Staphylococcus aureus[J]. Food Science, 2013, 34(16): 169−172. doi: 10.7506/spkx1002-6630-201316034
[38] GARRIDO-MAESTU A, AZINHEIRO S, CARVALHO J, et al. Rapid and sensitive detection of viable Listeria monocytogenes in food products by a filtration-based protocol and qPCR[J]. Food Microbiology,2018,73:254−263. doi: 10.1016/j.fm.2018.02.004
[39] 王建昌, 王金凤, 李静, 等. 基于内参的志贺氏菌实时荧光定量PCR快速检测[J]. 食品与生物技术学报,2016,35(4):408−418. [WANG Jianchang, WANG Jinfeng, LI Jing, et al. Establishment of fluorescence quantitative PCR assayin detection of Shigella based on internal reference[J]. Journal of Food Science and Biotechnology,2016,35(4):408−418.] doi: 10.3969/j.issn.1673-1689.2016.04.012 WANG Jianchang, WANG Jinfeng, LI Jing, et al. Establishment of fluorescence quantitative PCR assayin detection of Shigella based on internal reference[J]. Journal of Food Science and Biotechnology, 2016, 35(4): 408−418. doi: 10.3969/j.issn.1673-1689.2016.04.012
[40] 李丹丹, 徐义刚, 邱索平, 等. 金黄色葡萄球菌实时荧光PCR快速检测方法的建立[J]. 食品与发酵工业,2016,42(3):198−201. [LI Dandan, XU Yigang, QIU Suoping, et al. Development of a dual real-time PCR method for rapid detection of Staphylococcus aureus[J]. Food and Fermentation Industries,2016,42(3):198−201.] LI Dandan, XU Yigang, QIU Suoping, et al. Development of a dual real-time PCR method for rapid detection of Staphylococcus aureus[J]. Food and Fermentation Industries, 2016, 42(3): 198−201.
[41] 章沙沙. 实时荧光定量PCR检测食品中常见食源性致病菌[J]. 食品与发酵科技,2016,52(4):87−89. [ZHANG Shasha. Real-time fluorescent quantitative PCR for the detection of commonfood-borne pathogenic bacteria[J]. Food and Fermentation Sciences and Technology,2016,52(4):87−89.] ZHANG Shasha. Real-time fluorescent quantitative PCR for the detection of commonfood-borne pathogenic bacteria[J]. Food and Fermentation Sciences and Technology, 2016, 52(4): 87−89.
[42] 索原杰. 多重实时荧光PCR致病菌检测方法的构建及其在牛奶中的应用[D]. 杭州:浙江大学, 2018. [SUO Yuanjie. Developent of multiplex real-time PCR for pathogen detection and its application in milk[D]. Hangzhou:Zhejiang University, 2018.] SUO Yuanjie. Developent of multiplex real-time PCR for pathogen detection and its application in milk[D]. Hangzhou: Zhejiang University, 2018.
[43] 黄瑾, 梁涛波, 许恒毅. 数字PCR在生物学检测中应用的研究进展[J]. 生命科学,2021,33(2):255−264. [HUANG Jin, LIANG Taobo, XU Hengyi, et al. Research progress of application of digital PCR in biological detection[J]. Chinese Bulletin of Life Sciences,2021,33(2):255−264.] HUANG Jin, LIANG Taobo, XU Hengyi, et al. Research progress of application of digital PCR in biological detection[J]. Chinese Bulletin of Life Sciences, 2021, 33(2): 255−264.
[44] 刘可, 李梦霞, 张亮, 等. 数字PCR技术在食品检测中的应用研究进展[J]. 食品工业科技,2021,42(4):319−324. [LIU Ke, LI Mengxia, ZHANG Liang, et al. Research progress of digital PCR technology and its application in food detection[J]. Science and Technology of Food Industry,2021,42(4):319−324.] LIU Ke, LI Mengxia, ZHANG Liang, et al. Research progress of digital PCR technology and its application in food detection[J]. Science and Technology of Food Industry, 2021, 42(4): 319−324.
[45] NYARUABA R, MWALIKO C, KERING K K, et al. Droplet digital PCR applications in the tuberculosis world[J]. Tuberculosis,2019,117:85−92. doi: 10.1016/j.tube.2019.07.001
[46] BIAN X, JING F, LI G, et al. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes[J]. Biosensors and Bioelectronics,2015,74:770−777. doi: 10.1016/j.bios.2015.07.016
[47] LI H, BAI R, ZHAO Z, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases[J]. Bioscience Reports,2018,38(6):BSR20181170. doi: 10.1042/BSR20181170
[48] SUO Y, YIN W, ZHU Q, et al. A specific and sensitive aptamer-based digital PCR chip for Salmonella typhimurium detection[J]. Biosensors,2022,12(7):458. doi: 10.3390/bios12070458
[49] 程逸宇, 冯秋实, 吴海晶, 等. 微滴数字PCR定量检测乳制品中单核细胞增生李斯特氏菌[J]. 中国乳品工业,2019,47(12):37−40. [CHENG Yiyu, FENG Qiushi, WU Haijing, et al. Quantitative detection of Listeria monocytogenes in dairy productsby droplet digital PCR[J]. China Dairy Industry,2019,47(12):37−40.] CHENG Yiyu, FENG Qiushi, WU Haijing, et al. Quantitative detection of Listeria monocytogenes in dairy productsby droplet digital PCR[J]. China Dairy Industry, 2019, 47(12): 37−40.
[50] 翁文川, 管锦绣, 谢会, 等. 水产品中副溶血性弧菌PMA-dd PCR活菌定量检测方法的研究[J]. 现代食品科技,2019,35(6):273−279,190. [WENG Wenchuan, GUAN Jinxiu, XIE Hui, et al. Study on PMA-ddPCR method for quantitative detection of live Vibrio parahaemolyticus in aquatic products[J]. Modern Food Science and Technology,2019,35(6):273−279,190.] WENG Wenchuan, GUAN Jinxiu, XIE Hui, et al. Study on PMA-ddPCR method for quantitative detection of live Vibrio parahaemolyticus in aquatic products[J]. Modern Food Science and Technology, 2019, 35(6): 273−279,190.
[51] 张宜文, 赵海波, 吴红, 等. 数字PCR在食品安全检测中的应用研究进展[J]. 分析测试学报,2020,39(5):672−680. [ZHANG Yiwen, ZHAO Haibo, WU Hong, et al. Progress on application of digital PCR in food safety detection[J]. Journal of Instrumental Analysis,2020,39(5):672−680.] doi: 10.3969/j.issn.1004-4957.2020.05.018 ZHANG Yiwen, ZHAO Haibo, WU Hong, et al. Progress on application of digital PCR in food safety detection[J]. Journal of Instrumental Analysis, 2020, 39(5): 672−680. doi: 10.3969/j.issn.1004-4957.2020.05.018
[52] WANG M, YANG J, GAI Z, et al. Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk[J]. International Journal of Food Microbiology,2018,266:251−256. doi: 10.1016/j.ijfoodmicro.2017.12.011
[53] 唐毓祎. 水产品弧菌的等温核酸扩增可视化检测方法的建立与应用[D]. 上海:上海海洋大学, 2017. [TANG Yuyi. Development and application of colorimetric detection of Vibrio in seafood based on isothermal amplification techniques[D]. Shanghai:Shanghai Ocean University, 2017.] TANG Yuyi. Development and application of colorimetric detection of Vibrio in seafood based on isothermal amplification techniques[D]. Shanghai: Shanghai Ocean University, 2017.
[54] YI M Y, LING L, NEOGI S B, et al. Real time loop-mediated isothermal amplification using a portable fluorescence scanner for rapid and simple detection of Vibrio parahaemolyticus[J]. Food Control,2014,41:91−95. doi: 10.1016/j.foodcont.2014.01.005
[55] 胡元庆, 黄玉萍, 李凤霞, 等. 水产品中副溶血性弧菌LAMP检测方法的优化[J]. 现代食品科技,2017,33(6):313−320,247. [HU Yuanqing, HUANG Yuping, LI Fengxia, et al. Optimization of loop-mediated isothermal amplification methods for the detection of Vibrio parahaemolyticus in aquatic products[J]. Modern Food Science and Technology,2017,33(6):313−320,247.] HU Yuanqing, HUANG Yuping, LI Fengxia, et al. Optimization of loop-mediated isothermal amplification methods for the detection of Vibrio parahaemolyticus in aquatic products[J]. Modern Food Science and Technology, 2017, 33(6): 313−320,247.
[56] 黄朱梁, 薛超波, 孙瑛, 等. 食品中单核细胞增生李斯特氏菌实时浊度(LAMP)检测方法的建立[J]. 食品工业,2015,36(1):277−280. [HUANG Zhuliang, XUE Chaobo, SUN Ying, et al. Development of a real-time turbidimeter-based loop-mediated isothermal amplification (LAMP) method for detection of Listeria monocytogenes[J]. The Food Industry,2015,36(1):277−280.] HUANG Zhuliang, XUE Chaobo, SUN Ying, et al. Development of a real-time turbidimeter-based loop-mediated isothermal amplification (LAMP) method for detection of Listeria monocytogenes[J]. The Food Industry, 2015, 36(1): 277−280.
[57] 关玉婷, 刘东, 常江, 等. 沙门氏菌可视化环介导等温扩增检测方法的建立及初步应用[J]. 食品科技,2019,44(11):357−361. [GUAN Yuting, LIU Dong, CHANG Jiang, et al. Visual loop-mediated isothermal amplification assay for the detection of Salmonella and preliminary detection in raw milk[J]. Food Science and Technology,2019,44(11):357−361.] GUAN Yuting, LIU Dong, CHANG Jiang, et al. Visual loop-mediated isothermal amplification assay for the detection of Salmonella and preliminary detection in raw milk[J]. Food Science and Technology, 2019, 44(11): 357−361.
[58] 纪懿芳, 胡文忠, 姜爱丽, 等. LAMP技术快速检测海产品副溶血弧菌的条件优化[J]. 食品工业科技,2015,36(20):59−63,67. [JI Yifang, HU Wenzhong, JIANG Aili, et al. Optimize of loop-mediated isothermal amplification technology indetection of Vibrio parahaemolyticus[J]. Science and Technology of Food Industry,2015,36(20):59−63,67.] JI Yifang, HU Wenzhong, JIANG Aili, et al. Optimize of loop-mediated isothermal amplification technology indetection of Vibrio parahaemolyticus[J]. Science and Technology of Food Industry, 2015, 36(20): 59−63,67.
[59] 纪懿芳, 胡文忠, 姜爱丽, 等. 海产品中副溶血弧菌的LAMP-HNB快速检测技术[J]. 食品与发酵工业,2015,41(7):142−148. [JI Yifang, HU Wenzhong, JIANG Aili, et al. Rapid diagnostic methods for Vibrio parahaemolyticus in seafood using LAMP-HNB[J]. Food and Fermentation Industries,2015,41(7):142−148.] JI Yifang, HU Wenzhong, JIANG Aili, et al. Rapid diagnostic methods for Vibrio parahaemolyticus in seafood using LAMP-HNB[J]. Food and Fermentation Industries, 2015, 41(7): 142−148.
[60] MARUYAMA F, KENZAKA T, YAMAGUCHI N, et al. Detection of bacteria carrying the stx2 gene by in situ loop-mediated isothermal amplification[J]. Applied and Environmental Microbiology,2003,69(8):5023−5028. doi: 10.1128/AEM.69.8.5023-5028.2003
[61] 白榕, 白琳琳, 汪少芸, 等. LAMP扩增产物检测方法研究进展及基因编辑技术在其中的应用[J]. 农业生物技术学报,2021,29(10):2016−2030. [BAI Rong, BAI Linlin, WANG Shaoyun, et al. Research progress of LAMP amplicons detection method and the application of gene editing technology[J]. Journal of Agricultural Biotechnology,2021,29(10):2016−2030.] BAI Rong, BAI Linlin, WANG Shaoyun, et al. Research progress of LAMP amplicons detection method and the application of gene editing technology[J]. Journal of Agricultural Biotechnology, 2021, 29(10): 2016−2030.
[62] 王富鑫. LAMP法检测副溶血性弧菌核酸及其防污染研究[D]. 青岛:青岛科技大学, 2018. [WANG Fuxin. Study on LAMP method to detect Vibrio parahaemolyticus and the control of carryover contamintion[D]. Qingdao:Qingdao University of Science and Technology, 2018.] WANG Fuxin. Study on LAMP method to detect Vibrio parahaemolyticus and the control of carryover contamintion[D]. Qingdao: Qingdao University of Science and Technology, 2018.
[63] 王米佳. 可视LAMP-荧光与浊度技术检测枸杞中金黄色葡萄球菌的研究[D]. 银川:宁夏大学, 2021. [WANG Mijia. Study on the detection of Staphylococcus aureus in wolfberry by visual LAMP-fluorescence and visual LAMP-turbidity technology[D]. Yinchuan:Ningxia University, 2021.] WANG Mijia. Study on the detection of Staphylococcus aureus in wolfberry by visual LAMP-fluorescence and visual LAMP-turbidity technology[D]. Yinchuan: Ningxia University, 2021.
[64] 李碧霞. LAMP实时浊度法快速检测肠出血性大肠杆菌O157的研究[D]. 广州:华南理工大学, 2014. [LI Bixia. A real-time turbidimeter-based loop-mediated isothermal amplification assay for rapid detection of E. coli O157[D]. Guangzhou:South China University of Technology, 2014.] LI Bixia. A real-time turbidimeter-based loop-mediated isothermal amplification assay for rapid detection of E. coli O157[D]. Guangzhou: South China University of Technology, 2014.
[65] CAO X, ZHAO L, ZHANG J, et al. Detection of viable but nonculturable Vibrio parahaemolyticus in shrimp samples using improved real-time PCR and real-time LAMP methods[J]. Food Control,2019,103:145−152. doi: 10.1016/j.foodcont.2019.04.003
[66] 田祥强, 王西, 邹作成, 等. 基于hipO基因的环介导等温核酸扩增技术检测食源性空肠弯曲菌[J]. 中国动物传染病学报,2023,31(5):114−119. [TIAN Xiangqiang, WANG Xi, ZOU Zuocheng, et al. Development of loop-mediated isothermal amplification based on hipO gene for detecting foodborne Campylobacter jejunum[J]. Chinese Journal of Animal Infectious Diseases,2023,31(5):114−119.] TIAN Xiangqiang, WANG Xi, ZOU Zuocheng, et al. Development of loop-mediated isothermal amplification based on hipO gene for detecting foodborne Campylobacter jejunum[J]. Chinese Journal of Animal Infectious Diseases, 2023, 31(5): 114−119.
[67] 陈琼, 董华夏, 戴小芳, 等. 畜禽肉中金黄色葡萄球菌活菌可视化LAMP检测方法的研究[J]. 食品工业科技,2020,41(20):235−240,245. [CHEN Qiong, DONG Huaxia, DAI Xiaofang, et al. Visualization LAMP detection method of living Staphylococcus aureus in livestock and poultry meat[J]. Science and Technology of Food Industry,2020,41(20):235−240,245.] CHEN Qiong, DONG Huaxia, DAI Xiaofang, et al. Visualization LAMP detection method of living Staphylococcus aureus in livestock and poultry meat[J]. Science and Technology of Food Industry, 2020, 41(20): 235−240,245.
[68] 姜朕元, 赵艳, 王婷, 等. FADV-4环介导等温扩增-HNB可视化检测方法的建立和应用[J]. 黑龙江畜牧兽医,2020(24):65−69,76,175. [JIANG Zhenyuan, ZHAO Yan, WANG Ting, et al. Establishment and application of visual loop-mediated isothermal amplification-HNB for FADV-4[J]. Heilongjiang Animal Science and Veterinary Medicine,2020(24):65−69,76,175.] JIANG Zhenyuan, ZHAO Yan, WANG Ting, et al. Establishment and application of visual loop-mediated isothermal amplification-HNB for FADV-4[J]. Heilongjiang Animal Science and Veterinary Medicine, 2020(24): 65−69,76,175.
[69] KUMAR Y. Isothermal amplification-based methods for assessment of microbiological safety and authenticity of meat and meat products[J]. Food Control,2021,121:107679. doi: 10.1016/j.foodcont.2020.107679
[70] ABDOLAHZADEH A, DOLGOSHEINA E V, UNRAU P J. RNA detection with high specificity and sensitivity using nested fluorogenic Mango NASBA[J]. RNA,2019,25(12):1806−1813. doi: 10.1261/rna.072629.119
[71] 杨蜜, 夏云. 核酸序列依赖扩增技术检测隐球菌的方法建立和评价[J]. 重庆医科大学学报,2020,45(6):833−837. [YANG Mi, XIA Yun. Development and evaluation of nucleic acid sequence-based amplification for detection of Cryptococcus neoformans[J]. Journal of Chongqing Medical University,2020,45(6):833−837.] YANG Mi, XIA Yun. Development and evaluation of nucleic acid sequence-based amplification for detection of Cryptococcus neoformans[J]. Journal of Chongqing Medical University, 2020, 45(6): 833−837.
[72] CLANCY E, COUGHLAN H, HIGGINS O, et al. Development of internally controlled duplex real-time NASBA diagnostics assays for the detection of microorganisms associated with bacterial meningitis[J]. Journal of Microbiological Methods,2016,127:197−202. doi: 10.1016/j.mimet.2016.06.017
[73] ZHAI L, LIU H, CHEN Q, et al. Development of a real-time nucleic acid sequence-based amplification assay for the rapid detection of Salmonella spp. from food[J]. Brazilian Journal of Microbiology,2019,50(1):255−261. doi: 10.1007/s42770-018-0002-9
[74] 倪鑫, 王志聪, 雷质文, 等. 依赖于核酸序列恒温扩增技术快速检测副溶血性弧菌方法的建立[J]. 中国预防兽医学报,2011,33(11):882−886. [NI Xin, WANG Zhicong, LEI Zhiwen, et al. Establishment of the nucleic acid sequence-based amplification method of detecting Vibrio parahaemolyticus[J]. Chinese Journal of Preventive Veterinary Medicine,2011,33(11):882−886.] doi: 10.3969/j.issn.1008-0589.2011.11.13 NI Xin, WANG Zhicong, LEI Zhiwen, et al. Establishment of the nucleic acid sequence-based amplification method of detecting Vibrio parahaemolyticus[J]. Chinese Journal of Preventive Veterinary Medicine, 2011, 33(11): 882−886. doi: 10.3969/j.issn.1008-0589.2011.11.13
[75] 陈盈竹. 基于核酸序列依赖扩增技术诊断侵袭性念珠菌病的实验研究[D]. 重庆:重庆医科大学, 2020. [CHEN Yingzhu. Reserch of nucleic acid sequence-based amplification assay for diagnosis of invasive candidlasis[D]. Chongqing:Chongqing Medical University, 2020.] CHEN Yingzhu. Reserch of nucleic acid sequence-based amplification assay for diagnosis of invasive candidlasis[D]. Chongqing: Chongqing Medical University, 2020.
[76] ABD E L WAHED A, PATEL P, FAYE O, et al. Recombinase polymerase amplification assay for rapid diagnostics of dengue infection[J]. PLoS One,2015,10(6):e0129682. doi: 10.1371/journal.pone.0129682
[77] 李凯. RPA等温扩增技术在转基因玉米检测中的应用[D]. 北京:中国农业科学院, 2017. [LI Kai. The application of RPA technology in detection of transgenic maize[D]. Beijing:Chinese Academy of Agricultural Sciences, 2017.] LI Kai. The application of RPA technology in detection of transgenic maize[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017.
[78] 贾玄. RPA等温扩增技术在转基因水稻检测中的应用[D]. 北京:中国农业科学院, 2017. [JIA Xuan. The application of recombinase polymerase amplification (RPA) technology in detection of transgenic rices[D]. Beijing:Chinese Academy of Agricultural Sciences, 2017.] JIA Xuan. The application of recombinase polymerase amplification (RPA) technology in detection of transgenic rices[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017.
[79] 洪晗露. 实时定量RPA技术用于婴儿配方奶粉中阪崎克罗诺杆菌和沙门氏菌的快速检测[D]. 广州:暨南大学, 2020. [HONG Hanlu. Real-time quantitative RPA technology for rapid detection Cronobacter sakazakii and Salmonella in powdered infant formula[D]. Guangzhou:Ji’nan University, 2020.] HONG Hanlu. Real-time quantitative RPA technology for rapid detection Cronobacter sakazakii and Salmonella in powdered infant formula[D]. Guangzhou: Ji’nan University, 2020.
[80] 刘立兵, 耿云云, 姜彦芬, 等. 沙门氏菌重组酶聚合酶检测方法的建立及应用[J]. 食品科学,2019,40(2):298−303. [LIU Libing, GENG Yunyun, JIANG Yanfen, et al. Development and application of a recombinase polymerase amplification assay for detection of Salmonella[J]. Food Science,2019,40(2):298−303.] doi: 10.7506/spkx1002-6630-20170906-094 LIU Libing, GENG Yunyun, JIANG Yanfen, et al. Development and application of a recombinase polymerase amplification assay for detection of Salmonella[J]. Food Science, 2019, 40(2): 298−303. doi: 10.7506/spkx1002-6630-20170906-094
[81] 刘立兵, 孙晓霞, 姜彦芬, 等. 食品中检测志贺氏菌的实时荧光RPA方法的建立与应用[J]. 中国食品学报,2019,19(10):259−264. [LIU Libing, SUN Xiaoxia, JIANG Yanfen, et al. Development and application of the real-time recombinase polymerase amplification assay for detection of Shigella in food[J]. Journal of Chinese Institute of Food Science and Technology,2019,19(10):259−264.] LIU Libing, SUN Xiaoxia, JIANG Yanfen, et al. Development and application of the real-time recombinase polymerase amplification assay for detection of Shigella in food[J]. Journal of Chinese Institute of Food Science and Technology, 2019, 19(10): 259−264.
[82] DU X, ZANG Y, LIU H, et al. Recombinase polymerase amplification combined with lateral flow strip for Listeria monocytogenes detection in food[J]. Journal of Food Science,2018,83(4):1041−1047. doi: 10.1111/1750-3841.14078
[83] 胡金强, 魏向珂, 黄润娜, 等. 食源性致病菌RPA检测技术研究进展[J]. 食品工业科技,2018,39(7):329−334. [HU Jinqiang, WEI Xiangke, HUANG Runna, et al. Advance in RPA detection technologies of foodborne pathogenic bacteria[J]. Science and Technology of Food Industry,2018,39(7):329−334.] HU Jinqiang, WEI Xiangke, HUANG Runna, et al. Advance in RPA detection technologies of foodborne pathogenic bacteria[J]. Science and Technology of Food Industry, 2018, 39(7): 329−334.
[84] 何洁, 朱超, 黄珊珊, 等. 食品中单核细胞增生李斯特菌的重组酶聚合酶扩增检测方法的建立[J]. 中国食品卫生杂志,2021,33(3):274−278. [HE Jie, ZHU Chao, HUANG Shanshan, et al. Development of recombinase polymerase amplification detection method for Listeria monocytogenes in food[J]. Chinese Journal of Food Hygiene,2021,33(3):274−278.] HE Jie, ZHU Chao, HUANG Shanshan, et al. Development of recombinase polymerase amplification detection method for Listeria monocytogenes in food[J]. Chinese Journal of Food Hygiene, 2021, 33(3): 274−278.
[85] 崔荣飞, 赵义良, 田梅, 等. 重组酶介导扩增技术快速检测食品中单核细胞增生李斯特菌[J]. 食品安全质量检测学报,2021,12(5):1773−1777. [CUI Rongfei, ZHAO Yiliang, TIAN Mei, et al. Rapid detection of Listeria monocytogenes by recombinase aided amplification method[J]. Journal of Food Safety and Quality,2021,12(5):1773−1777.] CUI Rongfei, ZHAO Yiliang, TIAN Mei, et al. Rapid detection of Listeria monocytogenes by recombinase aided amplification method[J]. Journal of Food Safety and Quality, 2021, 12(5): 1773−1777.
[86] XUE G, LI S, ZHAO H, et al. Use of a rapid recombinase-aided amplification assay for Mycoplasma pneumoniae detection[J]. BMC Infectious Diseases,2020,20(1):1−7. doi: 10.1186/s12879-019-4717-5
[87] ZHANG X, GUO L, MA R, et al. Rapid detection of Salmonella with recombinase aided amplification[J]. Journal of Microbiological Methods,2017,139:202−204. doi: 10.1016/j.mimet.2017.06.011
[88] 丁昕, 刘燕红, 倪碧娴, 等. 基于重组酶介导等温扩增技术的细粒棘球绦虫核酸检测方法的建立[J]. 中国血吸虫病防治杂志,2020,32(4):340−344. [DING Xin, LIU Yanhong, NI Bixian, et al. Establishment of a nucleic acid assay for detection of Echinococcus granulosus based on recombinase-aided isothermal amplification assay[J]. Chinese Journal of Schistosomiasis Control,2020,32(4):340−344.] DING Xin, LIU Yanhong, NI Bixian, et al. Establishment of a nucleic acid assay for detection of Echinococcus granulosus based on recombinase-aided isothermal amplification assay[J]. Chinese Journal of Schistosomiasis Control, 2020, 32(4): 340−344.
[89] 郝林慧, 梁莹, 罗纪军, 等. 重组酶介导扩增技术快速检测水产品中副溶血性弧菌[J]. 食品安全质量检测学报,2021,12(13):5266−5272. [HAO Linhui, LIANG Ying, LUO Jijun, et al. Rapid detection of Vibrio parahaemolyticus in aquatic products by recombinase aid amplification[J]. Journal of Food Safety and Quality,2021,12(13):5266−5272.] HAO Linhui, LIANG Ying, LUO Jijun, et al. Rapid detection of Vibrio parahaemolyticus in aquatic products by recombinase aid amplification[J]. Journal of Food Safety and Quality, 2021, 12(13): 5266−5272.
[90] 黄新新, 何宇平, 袁辰刚, 等. 荧光重组酶介导等温扩增检测食品中单增李斯特菌[J]. 微生物学通报,2021,48(12):4989−5000. [HUANG Xinxin, HE Yuping, YUAN Chengang, et al. Detection of Listeria monocytogenes in food by fluorescent recombinase-aided amplification[J]. Microbiology China,2021,48(12):4989−5000.] HUANG Xinxin, HE Yuping, YUAN Chengang, et al. Detection of Listeria monocytogenes in food by fluorescent recombinase-aided amplification[J]. Microbiology China, 2021, 48(12): 4989−5000.
[91] NASRABADI Z, RANJBAR R, POORALI F, et al. Detection of eight foodborne bacterial pathogens by oligonucleotide array hybridization[J]. Electronic Physician,2017,9(5):4405. doi: 10.19082/4405
[92] LI Y. Establishment and application of a visual DNA microarray for the detection of food-borne pathogens[J]. Analytical Sciences,2016,32(2):215−218. doi: 10.2116/analsci.32.215
[93] ASLAN H, EKINCI A, ASLAN İ. Nucleic acid-based methods in the detection of foodborne pathogens[M]. Natural Remedies for Pest, Disease and Weed Control. Academic Press, 2020:143−161.
[94] YIN H B, CHEN C H, KATCHMAN B, et al. Rapid detection of Salmonella enterica in leafy greens by a novel DNA microarray-based pathogenDX system[J]. Food Microbiology,2022,107:104086. doi: 10.1016/j.fm.2022.104086
[95] DAY J B, HAMMACK T S. Bio-plex suspension array immuno-detection of Listeria monocytogenes from cantaloupe and packaged salad using virulence protein inducing activated charcoal enrichment media[J]. Food Microbiology,2019,84:103225. doi: 10.1016/j.fm.2019.05.009
[96] HU W, FENG K, JIANG A, et al. An in situ-synthesized gene chip for the detection of food-borne pathogens on fresh-cut cantaloupe and lettuce[J]. Frontiers in Microbiology,2020,10:3089. doi: 10.3389/fmicb.2019.03089
[97] ZHAO X, WEI C, ZHONG J, et al. Research advance in rapid detection of foodborne Staphylococcus aureus[J]. Biotechnology & Biotechnological Equipment,2016,30(5):827−833.
[98] WANG X, YING S, WEI X, et al. Development of a gold nanoparticle-based universal oligonucleotide microarray for multiplex and low-cost detection of foodborne pathogens[J]. International Journal of Food Microbiology,2017,253:66−74. doi: 10.1016/j.ijfoodmicro.2017.05.005
-
期刊类型引用(7)
1. 周贯旭,姜红,徐雪芳. 光谱技术结合化学计量学在食源性致病菌检测中的研究进展. 化学通报. 2025(01): 83-92+82 .  百度学术
百度学术
2. 董雪,姜利,岳涛,王鹏,汤有宏. MALDI-TOF MS技术识别白酒酿造微生物的可行性探讨. 酿酒. 2025(01): 132-135 .  百度学术
百度学术
3. 张蕾,陈亮,冯万宇,苗艳,兰世捷,吴宪,沈思思,张艳,田秋丰,杨昊天,姚美玲,南景东,江波涛,史同瑞,黄宣凯. 多杀性巴氏杆菌重组酶聚合酶扩增快速诊断方法的建立. 中国兽医科学. 2025(01): 56-64 .  百度学术
百度学术
4. 冯茹荔,田晓青,马赛,杜建森. 基于核酸扩增技术的肠球菌快速检测方法研究进展. 中国国境卫生检疫杂志. 2025(01): 92-97 .  百度学术
百度学术
5. 张捷,陈勃旭,杜海宽,张子仑,严陶陶,陈佳,周巍. Taq Man探针实时荧光定量聚合酶链式反应检测低温酸乳中常见的霉菌和酵母. 乳业科学与技术. 2024(05): 20-24 .  百度学术
百度学术
6. 张涛,张晨,邢军梅,蔡宏芳,王会宝. 基因编辑技术在食源性微生物检测中的应用研究进展. 食品安全导刊. 2024(34): 119-121+125 .  百度学术
百度学术
7. 吕珍. 食品中食源性致病菌的检测与防控技术研究. 中外食品工业. 2024(19): 49-51 .  百度学术
百度学术
其他类型引用(1)





 下载:
下载:
 下载:
下载:



