High-throughput Sequencing to Analyze Changes in the Structural Diversity of the Flora of Cheddar Cheese during Processing
-
摘要: 为明确契达奶酪加工过程中的菌群结构,本研究采用高通量测序技术分析契达奶酪在加工过程中(巴氏杀菌后、凝乳和成熟0、30、60和90 d)三个阶段的菌群结构。结果表明,契达奶酪加工过程中各阶段微生物群落结构差异较大。巴氏杀菌后,微生物群落多样性和丰度均为最高(Chao1和Shannon指数平均值分别为6.09和1415.78);在属水平上,巴氏杀菌后的优势菌群为寡养单胞菌属(Stenotrophomonas,21.04%),在凝乳和成熟阶段菌群结构比较相似,乳球菌属(Lactococcus)为两阶段的绝对优势菌群,丰度平均值达85%以上;在成熟期内,乳球菌属的相对丰度呈先上升后下降的趋势;巴氏杀菌后的奶酪体系中菌群结构与其他组相比差异较大,并且随着成熟期的延长,各组菌群结构变化较小。该研究为明确契达奶酪菌群结构提供依据,对契达奶酪的微生物组信息扩充具有参考价值。Abstract: In order to clarify the microflora structure in Cheddar cheese processing, MiSeq high-throughput sequencing technology was used to analyze the community structure of Cheddar cheese at three stages of processing (post-pasteurization, curdling, and ripening 0, 30, 60 and 90 d) in this study. The results showed that the community structure varies widely of cheddar cheese during processing. The highest microbial community diversity and abundance were found after pasteurization (Chao1 index and Shannon index mean values were 6.09 and 1415.78, respectively). The dominant microflora in the pasteurization stage at the genus level was Stenotrophomonas (21.04%). The community structure was relatively similar in the curd and ripening stages, Lactococcus were the dominant flora in both stages, with abundance averaging more than 85%. During the ripening period, the relative abundance of Lactococcus increased first and then decreased. The community structure in the pasteurized cheeses was different compared to the other groups, and there was less change in the community structure of the groups during the ripening period. This study provides a basis for clarifying the community structure of Cheddar cheese, and has a certain reference value for the expansion of Cheddar cheese microbiome information.
-
Keywords:
- Cheddar cheese /
- process /
- high-throughput sequencing /
- flora structure
-
奶酪是一种含有丰富蛋白质、脂肪及全部必需氨基酸、多种维生素和矿物质的高营养乳制品。国际上将其划分为天然奶酪、再制奶酪和奶酪食品三大类。其中契达奶酪口味是最为温和清淡的一种天然奶酪品种,备受我国消费者喜爱,也是我国进口数量最多的奶酪品种[1]。
契达奶酪的加工过程主要包括原料乳巴氏杀菌、发酵、凝乳、排乳清、契达化和加盐等。此外,契达奶酪是一种成熟型奶酪,需要在一定温度和湿度条件下进行较长时间的成熟才能形成成品。其成熟过程涉及到一系列的化学反应,包括糖酵解、脂解分解和蛋白质水解等一系列的生物化学和微生物学反应,所以对奶酪加工和成熟过程中菌群结构进行测定是必要的。Zhang等[2]综合代谢组学和高通量测序对内蒙古传统奶酪加工过程中微生物群落演替与风味代谢物之间的动态相关性进行研究,发现其加工过程中微生物群落结构复杂影响着其风味物质的形成。Penland等[3]对P´elardon奶酪成熟过程中不同时期的样品微生物多样性进行研究,发现副干酪乳杆菌(Lactobacillus paracasei )、粪肠球菌(Enterococcus faecalis)和青霉菌(Penicillium)等微生物为优势菌群。Hao等[4]发现在契达奶酪成熟过程中添加益生菌更有可能促进更多生物活性肽产生,比如:血管紧张酶抑制剂(Angiotensin-converting enzyme inhibitory)。Tamang等[5]对奶酪等发酵乳制品中的微生物的功能特性进行总结综述,阐明了其对人体健康的积极作用。由此可见,在奶酪的加工和成熟过程中,体系中微生物参与多种生化代谢反应,产生多样化的代谢产物,影响奶酪的最终品质与营养[6]。
传统的奶酪微生物研究通常采用分离培养等方法,如变性梯度凝胶电泳[7](Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis,PCR-DGGE)和时相温度梯度凝胶电泳[8](Polymerase Chain Reaction-Temporal Temperature Gradient Electrophoresis,PCR-TTGE)等,对于传统发酵食品多菌种的复杂体系,难以快速、全面地分析微生物群落的真实结构。Hermet等[9]采用SSCP分析了36种奶酪表面微生物,发现了传统的分子生物学技术对奶酪整体微生物群落分析鉴定的局限。高通量测序技术(High-throughput sequencing)即下一代测序技术,可一次性对样品中的几十万到几百万条 DNA 分子进行序列测定[10]。它解决了PCR-TTGE、和PCR-DGGE等传统方法的局限[11],可快速确定样品中微生物的种类和丰度,具有分析全面、灵敏、快速等特点,能够更客观地反映样品中微生物群落结构的真实情况[12−13]。近年来,高通量测序技术已广泛应用于传统发酵食品的微生物群落的宏基因组研究中[14],Silvetti等[15]利用高通量测序技术对Silter 干酪成熟过程中的菌群结构进行测定。Camargo等[16]利用高通量测序技术对“Entre Serras”手工奶酪成熟过程中的微生物变化进行监测。高通量测序技术在探索微生物群落方面的广泛应用大大增加了人们对奶酪等传统发酵食品的认知[17],目前主要的研究方向集中在外源添加益生菌对奶酪菌群结构的影响,例如,Hanlon等[18]研究了在契达干酪制作过程中添加益生元,对其体系内菌群结构以及理化性质的影响。Vinderola等[19]将不同益生菌株加入乳制品中,对乳酸菌和益生菌之间的相互作用进行了探究,但目前利用高通量测序对无外源物质添加的天然奶酪在加工和成熟过程中的研究较少。
本文采用高通量测序技术监控契达奶酪加工过程中巴氏杀菌后、凝乳以及成熟三个阶段中微生物菌群结构的变化及其演替规律,其中成熟阶段周期较长,故在成熟期内分别选取成熟0、30、60、90 d共4组样本进行微生物复杂度分析,并从不同分类水平下分析加工过程中三个阶段契达奶酪的优势菌群,以期为功能性奶酪的开发提供理论基础。
1. 材料与方法
1.1 材料与设备
生牛乳 黑龙江龙丹乳业科技股份有限公司;氯化钙 食品级,天津市凯通化学试剂有限公司;凝乳酶 山东悠乐滋生物科技有限公司;商业发酵剂乳酸乳球菌乳酸亚种(Lactococcus lactis subsp. lactis)、乳酸乳球菌乳脂亚种(Lactococcus lactis subsp. cremoris) 美国科汉森公司;Power Soil DNA分离试剂盒 美国AXYGEN公司。
SYQ-DSX-280B手提式高压灭菌锅 上海申安医疗器械厂;CH-35干酪槽 黑龙江赫益乳业科技有限公司;HYCV-6小型干酪压榨器 黑龙江赫益乳业科技有限公司;14890真空包装机 得力集团有限公司。
1.2 实验方法
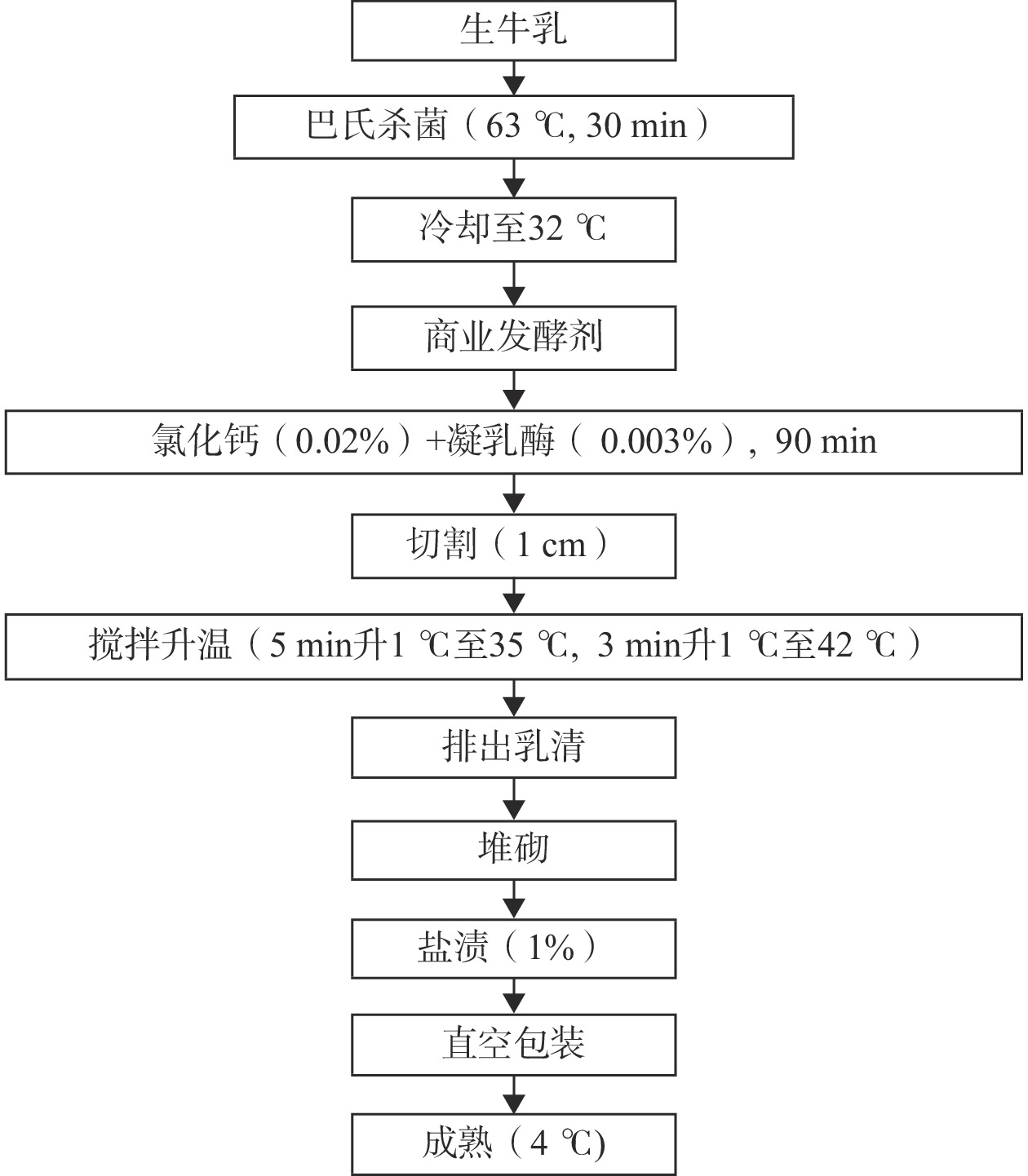
1.2.1 契达奶酪的加工
契达干酪的加工参照Hannon等[20]的方法并部分修改,具体加工过程如图1所示。添加0.01%(w/w)乳酸乳球菌乳酸亚种(Lactococcus lactis subsp. lactis)和乳酸乳球菌乳脂亚种(Lactococcus lactis subsp. cremoris)。选用巴氏杀菌后(F_milk)、凝乳阶段(Curd)、成熟阶段(Ch_0、Ch_30、Ch_60、Ch_90分别代表成熟0、30、60、90 d)的契达干酪样本,编号A、B、C、D、E、F共6组,每组样本取五份,共计30份样本。
1.2.2 DNA的提取
对上述六组契达奶酪进行DNA的提取,使用Power Soil DNA分离试剂盒,按照说明书提取。用1%琼脂糖凝胶电泳和分光光度法进行DNA质量和浓度检测。质检合格的样本储存在−20 ℃以备后续试验使用。
1.2.3 PCR 扩增和测序
首先将契达奶酪的DNA用特异引物对16S rDNA的V3-V4[21]进行扩增。引物序列为:338F(ACTCCTACGGGAGGCAGCAG)和806R(GGACTACHVGGGTWTCTAAT)。在上游和下游引物的5’末端各添加8 bp条形码序列,以区分不同的样本。PCR的扩增体系为25 μL:5 μL DNA(加入的DNA总量为30 ng)、1 μL Forward Primer(5 μmol/L)、1 μL Reverse Primer(5 μmol/L)、3 μL BSA 试剂、12.5 μL 2×Taq Plus Master Mix(DNA 聚合酶、2 ng/μL),加无菌双蒸水定容到25 μL。
在PCR仪上于94 ℃预变性5 min,使模板DNA充分变性,然后进入扩增循环。在每一个循环中,先于94 ℃保持30 s使模板变性,然后将温度降到50 ℃,保持30 s,使引物与模板充分退火;在72 ℃保持1 min,使引物在模板上延伸,合成DNA,完成一个循环。重复循环30次,使扩增的DNA片段大量累积。最后,在72 ℃保持7 min,使产物延伸完整,4 ℃保存。
对PCR扩增产物进行纯化、回收及定量处理。然后接测序接头,构建Miseq文库,最后上机测序。本试验委托北京奥维森基因科技有限公司进行高通量测序及原始数据的初级处理。PCR产物用于构建微生物多样性测序文库,用Illumina Miseq PE300高通量测序平台进行测序,测序原始序列上传至NCBI数据中心建立的SRA(Sequence Read Archive,http://www.ncbi.nlm.nih.gov/Traces/sra)数据库。
1.3 数据处理
高通量测序完成后,进行数据分析和序列优化[22]。下机数据经过QIIME1(v1.8.0)软件[23],根据Barcode序列拆分样本,使用Pear(v0.9.6)软件对数据进行过滤、拼接。去除打分低值于20,含有模糊碱基,引物错配序列。拼接后使用Vsearch(v2.7.1)软件去除长度小于230 bp的序列,并根据Gold Database数据库用uchime方法比对去除嵌合体序列。为了确保所有样本的覆盖率都相当高,把所有样本的数据量均一化至XXXX条序列。利用Qiime和Vsearch(v2.7.1),将OTU(97%相似水平下)进行生物信息统计分析。
2. 结果与分析
2.1 契达奶酪微生物群落Alpha多样性分析
Alpha反映单个奶酪样品内部物种的多样性,如表1所示,本实验中所有奶酪样品的Coverage ≥0.986,表明本实验的测序数据能够覆盖样品中的微生物种类,可以较为真实地反映奶酪样品中物种丰度及多样性,测序结果的有效性较高[24]。图2所示,稀释性曲线随着测序深度的增加而逐渐趋于平坦,表明本实验测序深度较好,测序数据量合理,能够反映契达奶酪的微生物群落组成[25]。
表 1 契达奶酪微生物群落丰度和多样性Table 1. Microbial community abundance and diversity of Cheddar cheese组别 样本 Coverage Shannon Simpson Chaol A组 F-milk-1 0.992 8.517 0.991 1599.573 F-milk-2 0.987 5.2 0.931 1240.551 F-milk-3 0.988 4.891 0.904 1277.736 F-milk-4 0.986 5.195 0.909 1438.525 F-milk-5 0.993 6.642 0.962 1522.493 B组 Curd-1 0.9896 0.255 0.203 1054.526 Curd-2 0.991 1.367 0.22 815.368 Curd-3 0.994 1.231 0.261 557.246 Curd-4 0.992 1.475 0.275 764.137 Curd-5 0.993 1.377 0.136 643.962 C组 Ch0-1 0.995 0.809 0.036 500.637 Ch0-2 0.995 0.688 0.114 568.529 Ch0-3 0.995 0.382 0.062 577.903 Ch0-4 0.995 0.557 0.104 506 Ch0-5 0.994 0.816 0.125 555.375 D组 Ch30-1 0.993 1.403 0.219 733.538 Ch30-2 0.994 0.737 0.115 622.3 Ch30-3 0.995 0.565 0.096 436.716 Ch30-4 0.996 0.567 0.106 353.171 Ch30-5 0.995 1.004 0.174 519.521 E组 Ch60-1 0.992 0.768 0.116 740.5 Ch60-2 0.994 0.714 0.116 584.662 Ch60-3 0.995 0.254 0.035 479.122 Ch60-4 0.994 0.515 0.078 545.838 Ch60-5 0.992 0.498 0.077 803.5 F组 Ch90-1 0.995 0.357 0.053 577 Ch90-2 0.992 0.821 0.124 711.02 Ch90-3 0.993 1.152 0.178 667.233 Ch90-4 0.992 0.839 0.128 879.55 Ch90-5 0.993 0.501 0.083 691.077 采用α多样性指标的Shannon 指数、Simpson指数与Chao1指数分别对六组样品微生物物种丰富度和多样性进行评估。Shannon指数和Simpson指数反映该组微生物群落多样性和物种分布情况。其中,Shannon指数常用来评价微生物群落的物种多样性,对样品中的微生物群落物种丰富度更为敏感,指数越大代表样品中的微生物多样性越高,Chaol指数反映该组微生物丰度,指数越大代表样品中的微生物丰度越高[26]。契达奶酪在加工和成熟过程中微生物群落多样性和物种丰富度变化如表1所示。结果表明,A组内样品Chao1指数和Shannon指数的平值均最高,即微生物群落多样性和丰度平均值均为最高,多样性指数平均值为6.09±1.52,丰度指数平均值为1415.78±154.47,而在E组微生物群落多样性最低,指数平均值为0.55±0.20;C组微生物群落丰度最低,指数平均值为541.69±35.98。B组内样品Chao1指数差异大,变化范围为557.25~1054.53,平均值为767.05±189.84。成熟初期(D组)组内样品Chao1指数平均值为533.05±149.98,成熟中期(E组)和成熟后期(F组)Chao1指数平均值分别为630.72±136.21和705.18±110.12,物种丰度呈现上升的趋势,但上升幅度明显降低,表明成熟过程中样品内的微生物正在逐渐适应生长环境,丰度最终有趋于稳定的可能。成熟过程中,Shannon 指数平均值分别为0.86±0.35、0.55±0.20和 0.73±0.31,表明契达奶酪成熟过程微生物群落多样性呈先下降后上升的趋势。
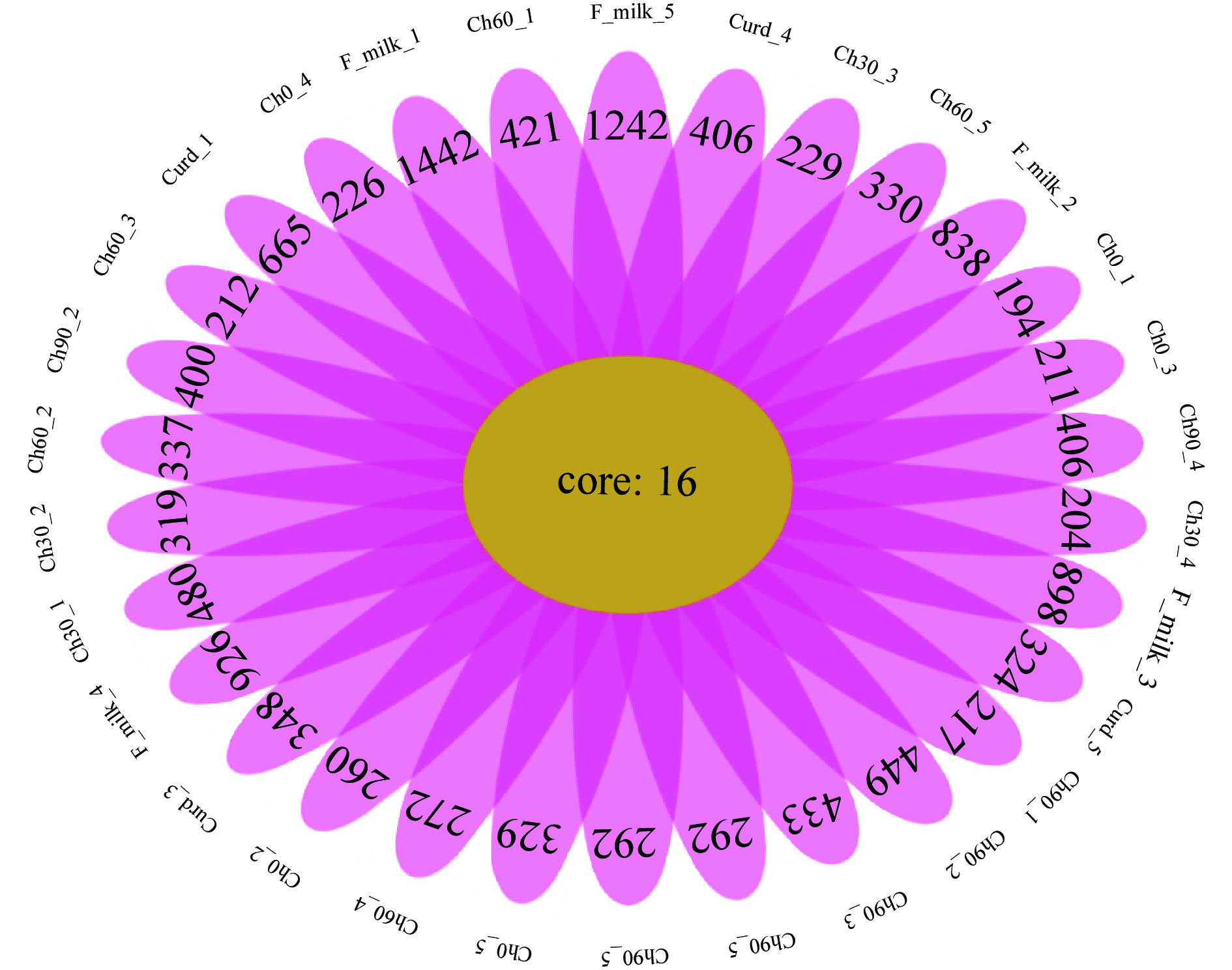
2.2 契达奶酪OTU分布的花瓣图分析
OTUs(Operational Taxonomic Units)是对奶酪样品中的序列进行归类操作(cluster),一般情况下,将相似性高于97%的不同 16S rDNA 序列定义为一个OTU,每一个OTU通常被视为一个微生物物种[27]。根据OTUs聚类分析结果绘制花瓣图(图3)。花瓣图是一种表示样本/组间特有和共有OTU数目的展示方式,每个花瓣代表一组样品,core数字代表每组共有OTU数目,每个花瓣上的数字代表该组特有的OTU数目。由图可知,契达奶酪样品共有OTU数量为16个,其中具体各组样品平均OTU数目分别为:巴氏杀菌后(F_milk):(1458个;854个;914个,942个;1258个)、凝乳阶段(Curd):(681个;465个;364个;422个;340个)、成熟0 d(Ch_0):(210个;276个;227个;242个;345个)、成熟30 d(Ch_30):(496个;335个;245个;220个;308个)、成熟60 d(Ch_60437个;353个;228个;288个;346个)、成熟90 d(Ch_90):(233个;416个;449个;422个;278个)。花瓣图结果表明:在所测样本内,巴氏杀菌后的菌群种类最为丰富,在凝乳阶段其菌群种类丰度逐渐下降,随着成熟时间的延长,奶酪体系内的菌群种类丰富度逐渐增加,到成熟末期,契达奶酪的特有OTU数大于成熟初期,说明成熟末期,契达奶酪体系中菌群种类较成熟初期更加丰富。
2.3 不同水平上契达奶酪菌群结构分析
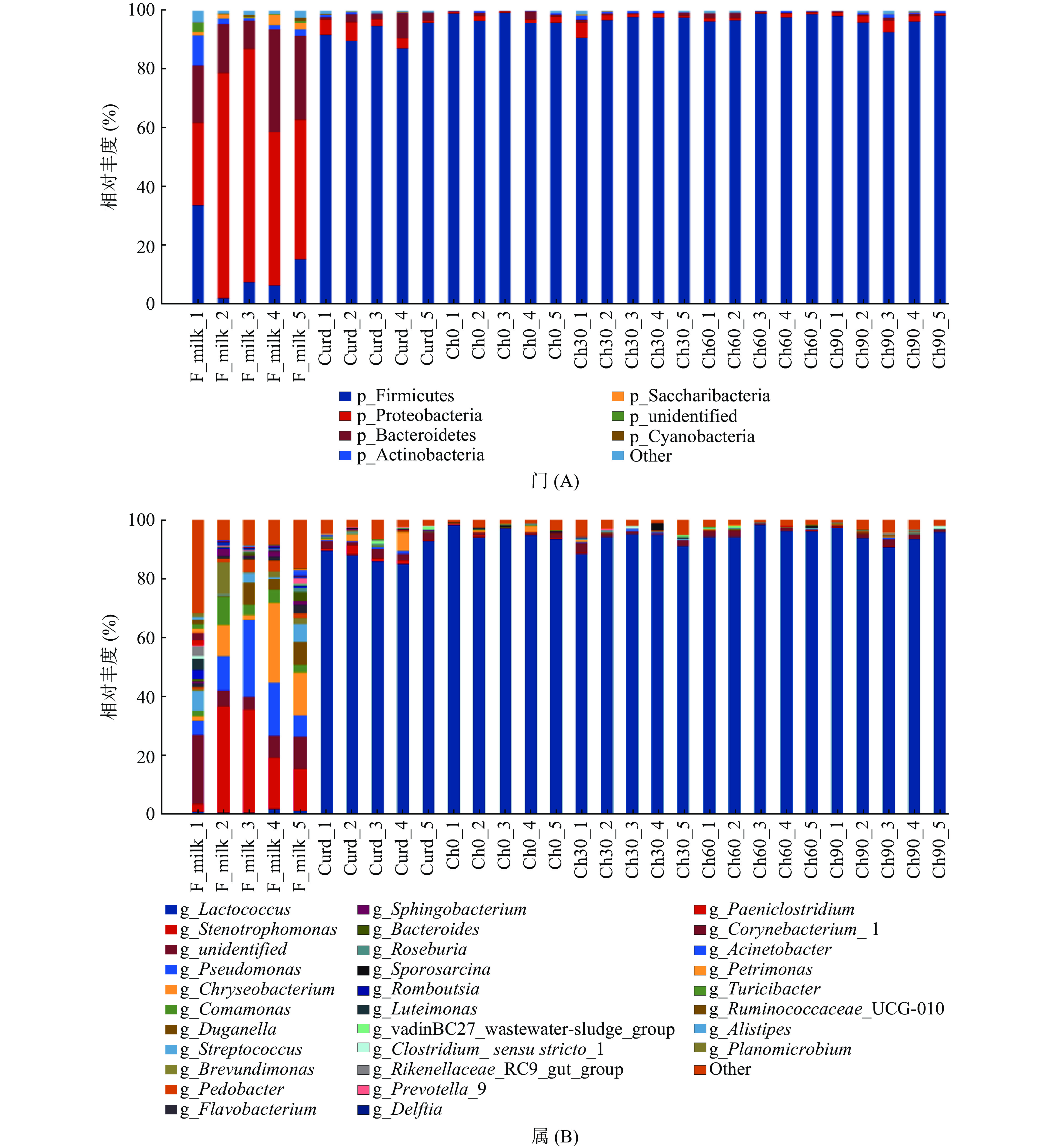
将各组契达奶酪样品中得到的OTUs序列从门和属2个水平进行物种注释,得到每个OTU在不同分类水平的物种分类信息,观测样本在不同分类水平上的群落结构。将多个样本的群落结构分析放在一起,观测其变化情况。用柱状图可视化观察不同分组的物种组成情况可以看到不同水平上物种的相对丰度,如图4所示。
在门水平对契达奶酪中的菌群结构进行分析,结果如图4A所示。变形菌门(Proteobacteria)、厚壁菌门(Firmicutes)和拟杆菌门(Bacteroidetes)为巴氏杀菌后样品的优势菌门,其相对丰度均值之和达到91.77%,其中变形菌门(Proteobacteria)为该组内的绝对优势菌门,其相对丰度均值达到56.80%,而在其余五组中变形菌门的相对丰度分别为3.58%、1.16%、1.90%、0.78%、0.20%,这可能是由于变形菌门的大部分细菌为好氧型,不能适应契达奶酪发酵过程中缺氧、产酸的环境,因而在发酵后的凝乳阶段以及成熟阶段其丰度迅速大幅度降低。除巴氏杀菌后期的样品以外,其余样品中厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)和变形菌门(Proteobacteria)均为各组样品中的优势菌门,其中厚壁菌门(Firmicutes)为契达奶酪成熟阶段的绝对优势菌门,成熟阶段四组样品相对丰度均值均达到96%以上,并且整个成熟过程中,其相对丰度变化不大趋于稳定。可见,契达奶酪制作过程中各阶段的优势菌门存在明显差异,巴氏杀菌后的优势菌门与原料奶中大体一致,主要由厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)和变形菌门(Proteobacteria)构成。然而在凝乳步骤之后,厚壁菌门(Firmicutes)则表现为绝对优势菌门,此外还有少量的变形菌门(Proteobacteria),这与Aldrete等[17]对墨西哥Poro奶酪制作过程中菌群结构的研究一致。
图4B显示了在属水平上对契达奶酪的菌群结构的分析结果,可以明显看出巴氏杀菌后契达奶酪体系内的菌属展现着高度的多样性,已鉴定出来的菌属就有20余种。而在凝乳和成熟两阶段中的多样性明显降低,这与张敏等[28]对乳制品微生物多样性的总结综述一致,这是由于原料奶发酵后的奶酪中,乳酸菌发酵乳糖产酸形成的生态位和资源竞争优势,将不利于其余大部分微生物的生长,使乳酸菌成为各种发酵乳品中的优势菌群。其中巴氏杀菌后契达奶酪中的优势菌群为金黄杆菌属(Chryseobacterium)和寡养单胞菌属(Stenotrophomonas),相对丰度均值分别为11.07%和21.04%。而在奶酪凝乳和成熟两阶段中,乳球菌属(Lactococcus)则为绝对优势菌群,并且在成熟期间,其相对丰度呈先上升后下降的趋势,丰度均值均达92%以上,这与曾椿淋[29]的研究结果一致。在巴氏杀菌后的契达奶酪中还发现了假单胞菌属(Pseudomonas)的存在,但在凝乳和成熟阶段中均未检测到,这可能是由于发酵后的高酸性环境不利于其生长繁殖。契达奶酪中存在的乳球菌属(Lactococcus)和部分酵母菌有着促进乳品发酵的功能,而假单胞菌属(Pseudomonas)的存在则会造成乳制品的腐败变质,给产品带来安全隐患[28]。此外,较高含量的乳球菌属(Lactococcus)的存在可能是由于在契达干酪发酵阶段加入了乳酸乳球菌乳酸亚种(Lactococcus lactis subsp. lactis)和乳酸乳球菌乳脂亚种(Lactococcus lactis subsp. cremoris),说明在奶酪中最主要的微生物是发酵剂微生物。Choi等[30]的研究也表明奶酪微生物群落的组成在很大程度上取决于所使用的发酵剂种类。但是在凝乳阶段和成熟阶段的样品中仍然发现了相对丰度较高的寡养单胞菌属(Stenotrophomonas),说明不能仅仅只关注奶酪中已知占优势的乳酸菌类发酵剂微生物,也要对这些之前未被关注但数量不少的其他微生物进行研究。
2.4 契达奶酪Beta多样性分析
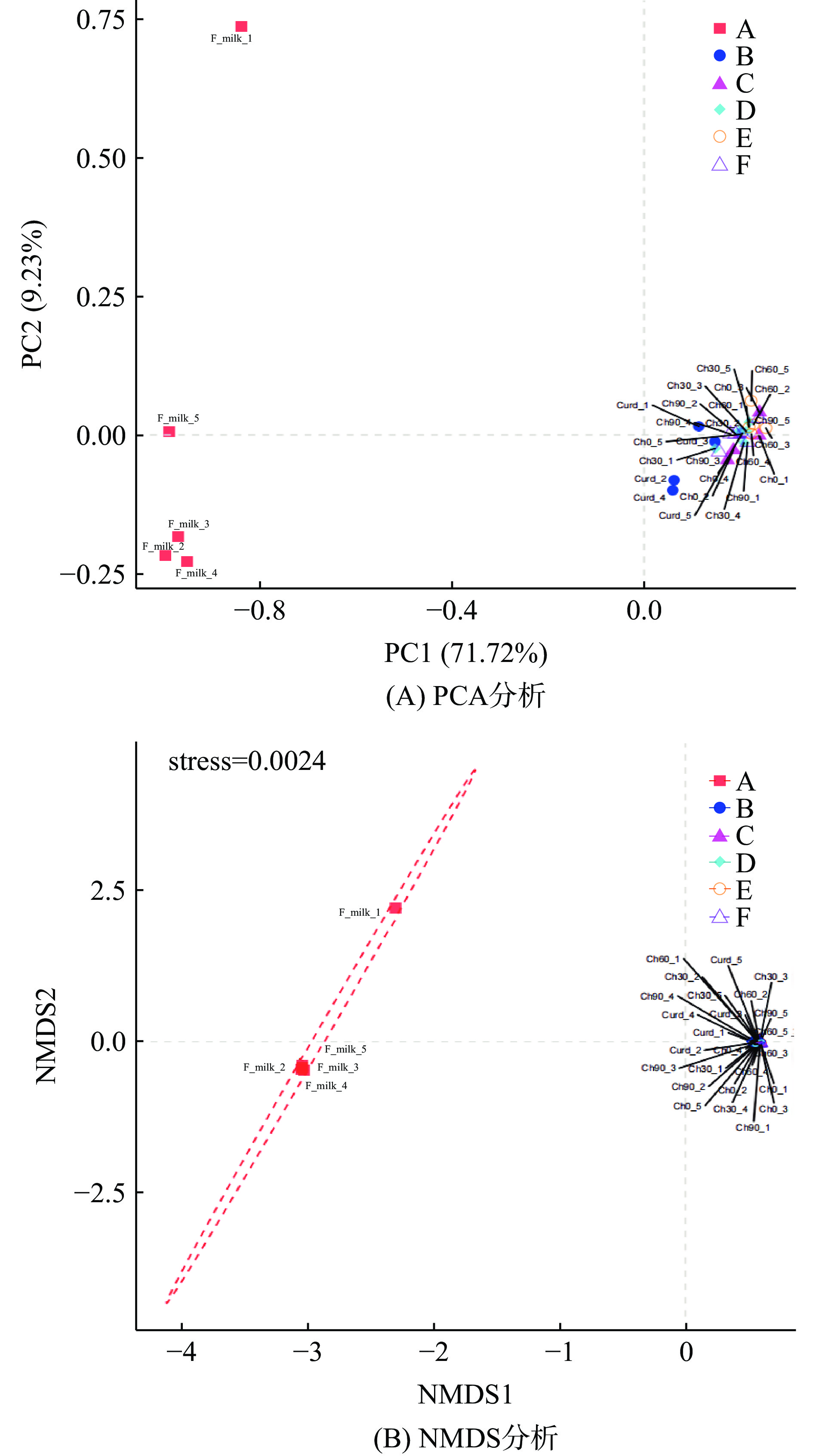
Beta多样性分析是对两组干酪样本微生物群落构成的差异。本研究采用PCA(Principal Component Analysis)分析和NMDS(Non-metric Multidimensional Scaling)两种方式分析比较契达奶酪加工阶段的样本群落构成差异。PCA分析,即主成分分析,通过降维处理,把多维数据作图,直观每组样本间群落构成的相似性[31]。NMDS分析同样是通过对原始数据的降维处理,研究不同组样本间群落构成的相似情况[32]。通过点与点的距离表现在图中,两点之间的距离越远,表明两个样品中微生物群落的差异越大[33],结果如图5A、B所示。
图5A所示,A组内五份样品分布较为分散,说明灭菌阶段的契达奶酪体系中菌群组成存在差异,而其余五组内样品分布较为集中,并且成熟阶段的四组样品分布十分集中,说明凝乳阶段和成熟阶段的契达奶酪体系中各组内菌群组成较相似,成熟阶段各组菌群结构存在高度相似。图5B所示,对群落测序结果进行排序分析,其stress值为0.0024<0.1,排序结果良好。A组与其它五组的契达干酪均不存在重合现象,且相距较远,说明灭菌阶段的奶酪体系中菌群结构与凝乳和成熟阶段相比差异较大,综合α多样性和菌群结构分析可知,此种差异主要来源于细菌相对丰度的改变,可能是由于灭菌后的原料乳在发酵过程中产生的酸类及醇类等物质的累积,不利于某些微生物的生长繁殖,引起微生物总体丰度下降,导致发酵后取样的凝乳阶段以及成熟阶段内菌群结构与灭菌阶段相比具有明显差异。另外,在不同成熟阶段各组样品之间都存在明显重叠部分,相距很近,说明成熟期的延长对其菌群结构影响较小,这可能是由于成熟过程中微生物已经逐渐适应了当前的生长环境,因此各组内菌群结构稳定而呈现明显的相似性。
3. 结论
本研究采用高通量测序分析契达奶酪加工过程中菌群变化情况,发现契达奶酪加工各阶段微生物群落结构差异较大。巴氏杀菌后的优势菌门为厚壁菌门、拟杆菌门和变形菌门,而在凝乳后,厚壁菌门则表现为绝对优势菌门,此外还存在少量的变形菌门。在属水平上,巴氏杀菌后契达奶酪体系内菌属体现出高度的多样性,而在凝乳和成熟两阶段多样性明显降低。巴氏杀菌后优势菌群为寡养单胞菌属,凝乳和成熟阶段菌群结构相似,乳球菌属为两阶段的绝对优势菌群。同时Beta多样性分析结果也表明巴氏杀菌后奶酪体系中的菌群结构与另外两阶段相比差异较大,成熟期的延长对菌群结构影响较小。本研究通过高通量测序技术监控契达奶酪加工过程中微生物菌群结构的变化,不仅可以锚定优势菌种,还将为契达奶酪体系内的微生态结构调控奠定理论基础,为探究添加外源物质对契达奶酪加工过程中原本菌群结构的影响提供对照参考,但本研究仅在属水平上对菌种进行鉴定,之后拟采用全基因组测序技术在种水平对其进一步分析。
-
表 1 契达奶酪微生物群落丰度和多样性
Table 1 Microbial community abundance and diversity of Cheddar cheese
组别 样本 Coverage Shannon Simpson Chaol A组 F-milk-1 0.992 8.517 0.991 1599.573 F-milk-2 0.987 5.2 0.931 1240.551 F-milk-3 0.988 4.891 0.904 1277.736 F-milk-4 0.986 5.195 0.909 1438.525 F-milk-5 0.993 6.642 0.962 1522.493 B组 Curd-1 0.9896 0.255 0.203 1054.526 Curd-2 0.991 1.367 0.22 815.368 Curd-3 0.994 1.231 0.261 557.246 Curd-4 0.992 1.475 0.275 764.137 Curd-5 0.993 1.377 0.136 643.962 C组 Ch0-1 0.995 0.809 0.036 500.637 Ch0-2 0.995 0.688 0.114 568.529 Ch0-3 0.995 0.382 0.062 577.903 Ch0-4 0.995 0.557 0.104 506 Ch0-5 0.994 0.816 0.125 555.375 D组 Ch30-1 0.993 1.403 0.219 733.538 Ch30-2 0.994 0.737 0.115 622.3 Ch30-3 0.995 0.565 0.096 436.716 Ch30-4 0.996 0.567 0.106 353.171 Ch30-5 0.995 1.004 0.174 519.521 E组 Ch60-1 0.992 0.768 0.116 740.5 Ch60-2 0.994 0.714 0.116 584.662 Ch60-3 0.995 0.254 0.035 479.122 Ch60-4 0.994 0.515 0.078 545.838 Ch60-5 0.992 0.498 0.077 803.5 F组 Ch90-1 0.995 0.357 0.053 577 Ch90-2 0.992 0.821 0.124 711.02 Ch90-3 0.993 1.152 0.178 667.233 Ch90-4 0.992 0.839 0.128 879.55 Ch90-5 0.993 0.501 0.083 691.077 -
[1] 干佳俪, 谭励, 王蓓, 等. 切达奶酪挥发性特征风味物质的多典型相关融合分析[J]. 中国乳品工业,2021,49(9):12−18,27. [GAN Jiali, TAN Li, WANG Bei, et al. Dentification of volatile flavor characteristics of Cheddar cheese with Multi-CCA Fusion[J]. China Dairy Industry,2021,49(9):12−18,27.] GAN Jiali, TAN Li, WANG Bei, et al . Dentification of volatile flavor characteristics of Cheddar cheese with Multi-CCA Fusion[J]. China Dairy Industry,2021 ,49 (9 ):12 −18,27 .[2] ZHANG X, ZHENG Y R, FENG J, et al. Integrated metabolomics and high-throughput sequencing to explore the dynamic correlations between flavor related metabolites and bacterial succession in the process of Mongolian cheese production[J]. Food Research International,2022,160:111672. doi: 10.1016/j.foodres.2022.111672
[3] PENLAND M, FALENTIN H, PARAYRE S, et al. Linking Pélardon artisanal goat cheese microbial communities to aroma compounds during cheese-making and ripening[J]. International Journal of Food Microbiology,2021,345:109130. doi: 10.1016/j.ijfoodmicro.2021.109130
[4] HAO Xinyue, XIA Yu , WANG Yuxin, et al. The addition of probiotic promotes the release of ACE-I peptide of Cheddar cheese:Peptide profile and molecular docking[J]. International Dairy Journal, 2023, 137:105507.
[5] TAMANG J P, SHIN D H, JUNG S J, et al. Functional properties of microorganisms in fermented foods[J]. Front Microbiol,2016,7:578.
[6] SAIDI V, SHEIKH-ZEINODDIN M, KOBARFARD F, et al. Profiling of bioactive metabolites during the ripening of a semi-hard non-starter culture cheese to detect functional dietary neurotransmitters[J]. Biocatalysis and Agricultural Biotechnology,2020,28:101734. doi: 10.1016/j.bcab.2020.101734
[7] DIMITRAKOPOULOU M E, PANTELELI E, VANTARAKIS A. Improved PCR-DGGE analysis by emulsion-PCR for the determination of food geographical origin:A case study on Greek PDO “avgotaracho Mesolonghiou”[J]. Current Research in Food Science,2021,4:746−751. doi: 10.1016/j.crfs.2021.10.005
[8] 董晓婉, 李宝坤, 李开雄, 等. 新疆蒙古族和哈萨克族传统乳制品中乳酸菌多样性的比较[J]. 食品工业科技,2013,34(21):162−166. [DONG Xiaowan, LI Baokun, LI Kaixiong, et al. Diversified comparison of lactic acid bacteria from traditional dairy products in Mongolian and Kazakh in Xinjiang[J]. Science and Technology of Food Industry,2013,34(21):162−166.] DONG Xiaowan, LI Baokun, LI Kaixiong, et al . Diversified comparison of lactic acid bacteria from traditional dairy products in Mongolian and Kazakh in Xinjiang[J]. Science and Technology of Food Industry,2013 ,34 (21 ):162 −166 .[9] HERMET A, MOUNIER J, KERAVEC M, et al. Application of capillary electrophoresis single-stranded conformation polymorphism (CE-SSCP) analysis for identification of fungal communities in cheese[J]. Food Microbiology,2014,41:82−90. doi: 10.1016/j.fm.2014.01.013
[10] SHENDURE J, BALASUBRAMANIAN S, CHURCH G M, et al. DNA sequencing at 40:past, present and future[J]. Nature,2017,550(7676):345−353. doi: 10.1038/nature24286
[11] WANG X H, WANG S H, ZHAO H. Unraveling microbial community diversity and succession of Chinese Sichuan sausages during spontaneous fermentation by high-throughput sequencing[J]. Journal of Food Science and Technology,2019,56(7):3254−3263. doi: 10.1007/s13197-019-03781-y
[12] YUN J M, ZHAO F Q, ZHANG W W, et al. Monitoring the microbial community succession and diversity of Liangzhou fumigated vinegar during solid-state fermentation with next-generation sequencing[J]. Annals of Microbiology,2019,69(3):279−289. doi: 10.1007/s13213-018-1418-z
[13] 熊香元, 张立钊, 陈力力, 等. 米粉发酵过程中乳酸菌多样性及功能分析[J]. 食品科学,2020,41(18):85−91. [XIONG Xiangyuan, ZHANG Lizhao, CHEN Lili, et al. Analysis of the diversity and function of lactic acid bacteria in fermented rice noodles during fermentation[J]. Food Science,2020,41(18):85−91.] XIONG Xiangyuan, ZHANG Lizhao, CHEN Lili, et al . Analysis of the diversity and function of lactic acid bacteria in fermented rice noodles during fermentation[J]. Food Science,2020 ,41 (18 ):85 −91 .[14] LIU A Q, YANG X, GUO Q Y, et al. Microbial communities and flavor compounds during the fermentation of traditional hong qu glutinous rice wine[J]. Foods,2022,11(8):1097. doi: 10.3390/foods11081097
[15] SILVETTI T, CAPRA E, MORANDI S, et al. Microbial population profile during ripening of Protected Designation of Origin (PDO) Silter cheese, produced with and without autochthonous starter culture[J]. LWT-Food Science and Technology,2017,84:821−831. doi: 10.1016/j.lwt.2017.06.022
[16] CAMARGO A C, COSTA E A, FUSIEGER A, et al. Microbial shifts through the ripening of the “Entre Serras” Minas artisanal cheese monitored by high-throughput sequencing[J]. Food Research International,2021,139:109803. doi: 10.1016/j.foodres.2020.109803
[17] ALDRETE T A, ESCOBAR R M C, TAMPLIN M L, et al. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese[J]. Food Microbiology,2014,44:136−141. doi: 10.1016/j.fm.2014.05.022
[18] HANLON M, CHOI J, GODDIK L et al. Microbial and chemical composition of Cheddar cheese supplemented with prebiotics from pasteurized milk to aging[J]. Journal of Dairy Science,2022,105(3):2058−2068. doi: 10.3168/jds.2021-21167
[19] VINDEROLA C G, MOCCHIUTTI P, REINHEIMER J A. Interactions among lactic acid starter and probiotic bacteria used for fermented dairy products[J]. Journal of Dairy Science,2002,85(4):721−729. doi: 10.3168/jds.S0022-0302(02)74129-5
[20] HANNON J A, WILKINSON M G, DELAHUNTY C M, et al. Use of autolytic starter systems to accelerate the ripening of Cheddar cheese[J]. International Dairy Journal,2003,13(4):313−323. doi: 10.1016/S0958-6946(02)00178-4
[21] CLAESSON M J, O'SULLIVAN O, WANG Q, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine[J]. PLoS One,2009,4(8):e6669. doi: 10.1371/journal.pone.0006669
[22] 吴树坤, 谢军, 卫春会, 等. 四川不同地区浓香型大曲微生物群落结构比较[J]. 食品科学,2019,40(14):144−152. [WU S K, XIE J, WEI C H, et al. Comparison of microbial structure of starter cultures (Daqu) for Luzhou flavor liquor in different regions of Sichuan[J]. Food Science,2019,40(14):144−152.] WU S K, XIE J, WEI C H, et al . Comparison of microbial structure of starter cultures (Daqu) for Luzhou flavor liquor in different regions of Sichuan[J]. Food Science,2019 ,40 (14 ):144 −152 .[23] BOLYEN E, RIDEOUT J R, DILLON M R, et al. Author correction:Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2[J]. Nat Biotechnol,2019,37(9):1091.
[24] 贾曼, 杨絮, 周国燕, 等. 高通量测序分析黄酒糟制香糟自然发酵过程中微生物群落多样性[J/OL]. 食品科学, 1−11 [2022-11-16]. http://kns.cnki.net/kcms/detail/11.2206.TS.20220819.0914.018.html. [JIA Man, YANG Xu, ZHOU Guoyan, et al. Microbial community diversity analysis during spontaneous fermentation of aromatic lees made from huang jiu (yellow rice wine) lees by high-throughput sequencin[J/OL]. Food Science, 1−11[2022-11-16].] JIA Man, YANG Xu, ZHOU Guoyan, et al. Microbial community diversity analysis during spontaneous fermentation of aromatic lees made from huang jiu (yellow rice wine) lees by high-throughput sequencin[J/OL]. Food Science, 1−11[2022-11-16].
[25] 王泽晗, 陶昱豪, 孙黎明, 等. 苗族酸酢鱼发酵过程中微生物群落、氨基酸及水分动态变化规律[J]. 食品科学,2021,42(12):116−122. [WANG Zehan, TAO Yuhao, SUN Liming, et al. Dynamic changes of microbial community, amino acid composition and moisture distribution during the fermentation of miao ethnic Suanzuo Fish[J]. Food Science,2021,42(12):116−122.] WANG Zehan, TAO Yuhao, SUN Liming, et al . Dynamic changes of microbial community, amino acid composition and moisture distribution during the fermentation of miao ethnic Suanzuo Fish[J]. Food Science,2021 ,42 (12 ):116 −122 .[26] ROSWELL M, DUSHOFF J, WINFREE R, et al. A conceptual guide to measuring species diversity[J]. Oikos,2021,130(3):321−338. doi: 10.1111/oik.07202
[27] 陈申习, 宿智新, 张磊, 等. 基于高通量测序的清香型和酱香型酒曲真菌群落特征研究[J]. 中国酿造,2021,40(7):5. [CHEN Shenxi, SU Zhixin, ZHANG Lei, et al. Fungal community characteristics of light-flavor and sauce-flavor Jiuqu based on high throughput sequencing[J]. China Brewing,2021,40(7):5.] CHEN Shenxi, SU Zhixin, ZHANG Lei, et al . Fungal community characteristics of light-flavor and sauce-flavor Jiuqu based on high throughput sequencing[J]. China Brewing,2021 ,40 (7 ):5 .[28] 张敏, 张艳, 兰国伟, 等. 高通量测序技术在乳制品微生物多样性中的研究进展[J]. 中国农学通报,2016,32(32):48−52. [ZHANG Min, ZHANG Yan, LAN Guowei, et al. Review of high-throughput sequencing technology in microbial diversity of dairy product[J]. Chinese Agricultural Science Bulletin,2016,32(32):48−52.] ZHANG Min, ZHANG Yan, LAN Guowei, et al . Review of high-throughput sequencing technology in microbial diversity of dairy product[J]. Chinese Agricultural Science Bulletin,2016 ,32 (32 ):48 −52 .[29] 曾椿淋. 内蒙古手工奶酪微生物多样性和功能性研究[D]. 南京:江苏大学, 2020. [ZENG Chunlin. Studies on the microbial diversity and functionality of microbes of inner mongolia handmade cheese[D]. Nanjing:Jiangsu University, 2020.] ZENG Chunlin. Studies on the microbial diversity and functionality of microbes of inner mongolia handmade cheese[D]. Nanjing: Jiangsu University, 2020.
[30] CHOI J, LEE S I, RACKERBY B, et al. Microbial communities of a variety of cheeses and comparison between core and rind region of cheeses[J]. Journal of Dairy Science,2020,103(5):4026−4042. doi: 10.3168/jds.2019-17455
[31] WANG Y, SHENG H F, HE Y, et al. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags[J]. Applied & Environmental Microbiology,2012,78(23):8264.
[32] NOVEAL R M, BURTON O T, WISE P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis[J]. Journal of Allergy and Clinical Immunology,2013,131(1):201−212. doi: 10.1016/j.jaci.2012.10.026
[33] 张敏, 张艳, 黄丽丽, 等. 基于16S rDNA高通量测序方法比较新疆西北部地区乳品中微生物的多样性[J]. 食品科学,2017,38(20):27−33. [ZHANG Min, ZHANG Yan, HUANG Lili, et al. 16S rDNA high-throughput sequencing-based methods to compare the microbial diversity in dairy products of the western and northern part of Xinjiang, China[J]. Food Science,2017,38(20):27−33.] ZHANG Min, ZHANG Yan, HUANG Lili, et al . 16S rDNA high-throughput sequencing-based methods to compare the microbial diversity in dairy products of the western and northern part of Xinjiang, China[J]. Food Science,2017 ,38 (20 ):27 −33 .





 下载:
下载:
 下载:
下载:



