| [1] |
KRUGER K, GRABOWSKI P J, ZAUG A J, et al. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena[J]. Cell,1982,31(1):147−157. doi: 10.1016/0092-8674(82)90414-7
|
| [2] |
GUERRIER-TAKADA C, GARDINER K, MARSH T, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme[J]. Cell,1983,35(3):849−857. doi: 10.1016/0092-8674(83)90117-4
|
| [3] |
ROBERTSOND L, JOYCE G F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA[J]. Nature,1990,344(6265):467−468. doi: 10.1038/344467a0
|
| [4] |
STRECKEROVA T, KURFURSTJ, CURTIS E A. Single-round deoxyribozyme discovery[J]. Nucleic Acids Research,2021,49(12):6971−6981. doi: 10.1093/nar/gkab504
|
| [5] |
PONCE S A, BOCCALETTO P, BUJNICKI J M. DNAmoreDB, a database of DNAzymes[J]. Nucleic Acids Research,2021,49(D1):D76−D81. doi: 10.1093/nar/gkaa867
|
| [6] |
BREAKER R R, JOYCE G F. A DNA enzyme that cleaves RNA[J]. Chemistry & Biology,1994,1(4):223−229.
|
| [7] |
ZHANG D P, WANG H I. Fluorescence anisotropy reduction of an allosteric G-rich oligonucleotide for specific silver ion and cysteine detection based on the G-Ag +-G base pair[J]. Analytical Chemistry,2019,91(22):14538−14544. doi: 10.1021/acs.analchem.9b03556
|
| [8] |
ZHU C, ZHAO X Y, YANG G, et al. Capillary electrophoresis involving in high efficiency screening for aptamers[J]. Chinese Journal of Analytical Chemistry,2020,48(5):583−589. doi: 10.1016/S1872-2040(20)60014-7
|
| [9] |
WANG H Y, LI X, LAI L A, et al. X-aptamers targeting Thy-1 membrane glycoprotein in pancreatic ductal adenocarcinoma[J]. Biochimie,2021,181:25−33. doi: 10.1016/j.biochi.2020.11.018
|
| [10] |
LIU H M, HAO J M, XU J, et al. Selection and identification of common aptamers against both Vibrio harveyi and Vibrio alginolyticus[J]. Chinese Journal of Analytical Chemistry,2020,48(5):623−631. doi: 10.1016/S1872-2040(20)60018-4
|
| [11] |
ZHENG Y, ZHAO Y W, DI Y, et al. DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer[J]. RSC Advances,2019,9(2):950−957. doi: 10.1039/C8RA08642G
|
| [12] |
SUN C Y, SU R F, BIE J X, et al. Label-free fluorescent sensor based on aptamer and thiazole orange for the detection of tetracycline[J]. Dyes and Pigments,2018,149:867−875. doi: 10.1016/j.dyepig.2017.11.031
|
| [13] |
WANG T, CHEN C, LARCHER L M, et al. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development[J]. Biotechnology Advances,2019,37(1):28−50. doi: 10.1016/j.biotechadv.2018.11.001
|
| [14] |
BAUM D A, SILVERMAN S K. Deoxyribozymes: Useful DNA catalysts in vitro and in vivo[J]. Cellular & Molecular Life Sciences Cmls,2008,65(14):2156−2174.
|
| [15] |
ROSENBACH H, BORGGRÄFE J, VICTOR J, et al. Influence of monovalent metal ions on metal binding and catalytic activity of the 10-23 DNAzyme[J]. Biological Chemistry,2021,402(1):99−111.
|
| [16] |
WANG Y, YANG F, YANG X. Label-free colorimetric biosensing of copper (II) ions with unimolecular self-cleaving deoxyribozymes and unmodified gold nanoparticle probes[J]. Nanotechnology,2010,21(20):205502. doi: 10.1088/0957-4484/21/20/205502
|
| [17] |
LEE Y, KLAUSER P C, BRANDSEN B M, et al. DNA-catalyzed DNA cleavage by a radical pathway with well-defined products[J]. Journal of the American Chemical Society,2017,139(1):255−261. doi: 10.1021/jacs.6b10274
|
| [18] |
GU H, FURUKAWA K, WEINBERG Z, et al. Small, highly active DNAs that hydrolyze DNA[J]. Journal of the American Chemical Society,2013,135(24):9121−9129. doi: 10.1021/ja403585e
|
| [19] |
BARLEV A, SEKHON G S, BENNET A J, et al. DNA repair by DNA: The UV1C DNA zyme catalyzes photoreactivation of cyclobutane thymine dimers in DNA more effectively than their de novo formation[J]. Biochemistry,2016,55(43):6010−6018. doi: 10.1021/acs.biochem.6b00951
|
| [20] |
BRANDSEN B M, HESSER A R, CASTNER M A, et al. DNA-catalyzed hydrolysis of esters and aromatic amides[J]. Journal of the American Chemical Society,2013,135(43):16014−16017. doi: 10.1021/ja4077233
|
| [21] |
ZHOU C, AVINS J L, KLAUSER P C, et al. DNA-catalyzed amide hydrolysis[J]. Journal of the American Chemical Society,2016,138(7):2106−2109. doi: 10.1021/jacs.5b12647
|
| [22] |
WANG Y M, SILVERMAN S K. Directing the outcome of deoxyribozyme selections to favor native 3'-5' RNA ligation[J]. Biochemistry,2005,44(8):3017−3023. doi: 10.1021/bi0478291
|
| [23] |
CUENOUD B, SZOSTAK J W. A DNA metalloenzyme with DNA ligase activity[J]. Nature,1995,375(6532):611−614. doi: 10.1038/375611a0
|
| [24] |
CHANDRA M, SILVERMAN S K. DNA and RNA can be equally efficient catalysts for carbon-carbon bond formation[J]. Journal of the American Chemical Society,2008,130(10):2936−2937. doi: 10.1021/ja7111965
|
| [25] |
WONG O Y, PRADEEPKUMAR P I, SILVERMAN S K. DNA-catalyzed covalent modification of amino acid side chains in tethered and free peptide substrates[J]. Biochemistry,2011,50(21):4741−4749. doi: 10.1021/bi200585n
|
| [26] |
CHU C C, WONG O Y, SILVERMAN S K. A generalizable DNA-catalyzed approach to peptide-nucleic acid conjugation[J]. ChemBioChem,2014,15(13):1905−1910. doi: 10.1002/cbic.201402255
|
| [27] |
LI Y F, BREAKER R R. Phosphorylating DNA with DNA[J]. Proceedings of the National Academy of Sciences,1999,96(6):2746−2751. doi: 10.1073/pnas.96.6.2746
|
| [28] |
MCMANUS S A, LI Y F. Multiple occurrences of an efficient self-phosphorylating deoxyribozyme motif[J]. Biochemistry,2007,46(8):2198−2204. doi: 10.1021/bi061613c
|
| [29] |
WALSH S M, SACHDEVA A, SILVERMAN S K. DNA catalysts with tyrosine kinase activity[J]. Journal of the American Chemical Society,2013,135(40):14928−14931. doi: 10.1021/ja407586u
|
| [30] |
CHANDRASEKAR J, WYLDER A C, SILVERMAN S K. Phosphoserine lyase deoxyribozymes: DNA-catalyzed formation of dehydroalanine residues in peptides[J]. Journal of the American Chemical Society,2015,137(30):9575−9578. doi: 10.1021/jacs.5b06308
|
| [31] |
LI Y F, LIU Y, BREAKER R R. Capping DNA with DNA[J]. Biochemistry,2000,39(11):3106−3114. doi: 10.1021/bi992710r
|
| [32] |
LI Y F, SEN D. Toward an efficient DNAzyme[J]. Biochemistry,1997,36(18):5589−5599. doi: 10.1021/bi962694n
|
| [33] |
YAO T J, PRZYBYLA J J, YEH P, et al. DNAzymes for amine and peptide lysine acylation[J]. Organic & Biomolecular Chemistry,2020,19(1):171−181.
|
| [34] |
HUANG P J J, LIU J. In vitro selection of chemically modified DNAzymes[J]. Chemistryopen,2020,9(10):1046−1059. doi: 10.1002/open.202000134
|
| [35] |
佟宗轩, 胡沁沁, 顾宏周. DNA酶: 筛选, 生物传感及展望[J]. 高等学校化学学报,2020,41(11):2345−2355. [TONG Z X, HU Q Q, GU H Z. Deoxyribozymes: Selection, biosensing and outlook[J]. Chemical Journal of Chinese Universities-Chinese,2020,41(11):2345−2355.
|
| [36] |
王月瑶. 催化RNA切割反应的新型短结合臂脱氧核酶[D]. 南京: 南京大学, 2019.
WANG Y Y. A novel small RNA-cleaving deoxyribozyme with a short binding arm[D]. Nanjing: Nanjing University, 2019.
|
| [37] |
SCHEITL C P M, LANGE S, HOBARTNER C. New deoxyribozymes for the native ligation of RNA[J]. Molecules,2020,25(16):3650. doi: 10.3390/molecules25163650
|
| [38] |
MORRISON D, ROTHENBROKER M, LI Y F. DNAzymes: Selected for applications[J]. Small Methods,2018,2(3):1700319. doi: 10.1002/smtd.201700319
|
| [39] |
SILVERMAN S K. Catalytic DNA: Scope, applications, and biochemistry of deoxyribozymes[J]. Trends in Biochemical Sciences,2016,41(7):595−609. doi: 10.1016/j.tibs.2016.04.010
|
| [40] |
ZHANG X B, KONG R M, LU Y. Metal ion sensors based on DNAzymes and related DNA molecules[J]. Annual Review of Analytical Chemistry,2011,4:105−128. doi: 10.1146/annurev.anchem.111808.073617
|
| [41] |
TORABI S F, WU P, MCGHEE C E, et al. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing[J]. Proceedings of the National Academy of Sciences,2015,112(19):5903−5908. doi: 10.1073/pnas.1420361112
|
| [42] |
LI H, HUANG X X, KONG D M, et al. Ultrasensitive, high temperature and ionic strength variation-tolerant Cu 2+ fluorescent sensor based on reconstructed Cu 2+-dependent DNAzyme/substratecomplex[J]. Biosensors and Bioelectronics,2013,42:225−228. doi: 10.1016/j.bios.2012.10.070
|
| [43] |
LI H, ZHANG Q, CAI Y, et al. Single-stranded DNAzyme-based Pb 2+ fluorescent sensor that can work well over a wide temperature range[J]. Biosensors and Bioelectronics,2012,34(1):159−164. doi: 10.1016/j.bios.2012.01.037
|
| [44] |
MOON W J, LIU J. Interfacing catalytic DNA with nanomaterials[J]. Advanced Materials Interfaces,2020,7(21):2001017. doi: 10.1002/admi.202001017
|
| [45] |
ZHAO X H, KONG R M, ZHANG X B, et al. Graphene-DNAzyme based biosensor for amplified fluorescence “turn-on” detection of Pb 2+ with a high selectivity[J]. Analytical Chemistry,2011,83(13):5062−5066. doi: 10.1021/ac200843x
|
| [46] |
YANG Z L, LOH K Y, CHU Y T, et al. Optical control of metal ion probes in cells and zebrafish using highly selective DNAzymes conjugated to upconversion nanoparticles[J]. Journal of the American Chemical Society,2018,140(50):17656−17665. doi: 10.1021/jacs.8b09867
|
| [47] |
范思思, 程进, 冀斌, 等. 脱氧核酶在生物检测及基因治疗中的研究进展[J]. 科学通报,2019,64:1027−1036. [FAN S S, CHEN J, JI B, et al. DNAzymes in biological detection and gene therapy[J]. Chin Sci Bull,2019,64:1027−1036. doi: 10.1360/N972018-00874
|
| [48] |
FAN H, ZHAO Z, YAN G, et al. A smart DNAzyme-MnO 2 nanosystem for efficient gene silencing[J]. Angewandte Chemie,2015,127(16):4883−4887. doi: 10.1002/ange.201411417
|
| [49] |
ZHANG J H, MA R, BLANCHARD A, et al. Conditional deoxyribozyme-nanoparticle conjugates for miRNA-triggered gene regulation[J]. ACS Applied Materials & Interfaces,2020,12(34):37851−37861.
|
| [50] |
李一凡, 吴燃峰, 杨静, 等. 基于DNA核酶的分子加密系统[J]. 信息网络安全,2017(6):43−48. [LI YF, WU R F, YANG J, et al. A molecule encryption system based on DNAzyme[J]. Netinfo Security,2017(6):43−48. doi: 10.3969/j.issn.1671-1122.2017.06.007
|
| [51] |
彭维平, 程丹华, 宋成. 基于多碱基组合映射编码和DNA计算的一次一密算法[J]. 计算机应用研究,2019,36(7):2190−2194. [PENG W P, CHENG D, SONG C. A one-cipher algorithm based on multi-base combination mapping coding and DNA calculation[J]. Application Research of Computers,2019,36(7):2190−2194. doi: 10.19734/j.issn.1001-3695.2018.01.0100
|





 本站查看
本站查看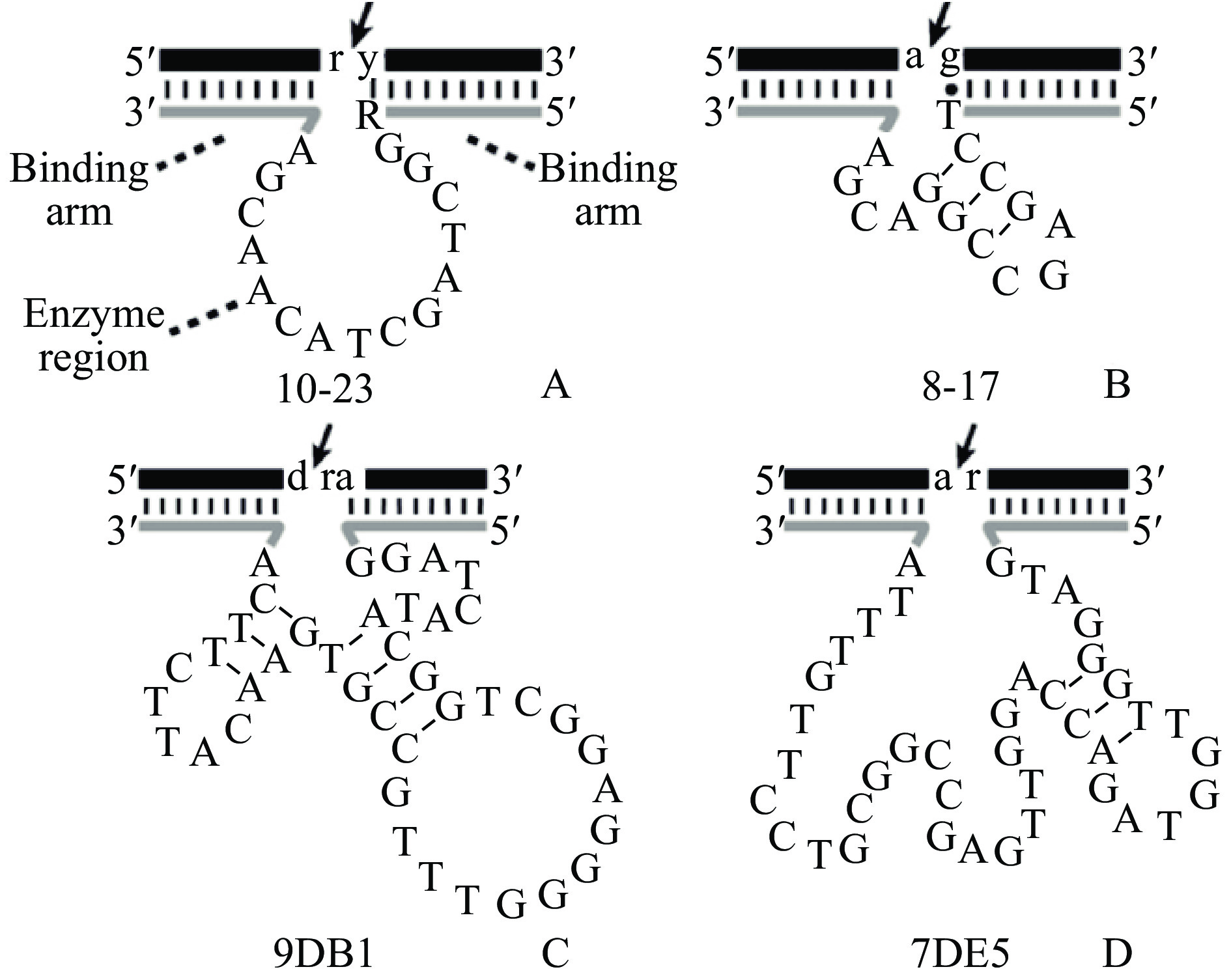




 DownLoad:
DownLoad: